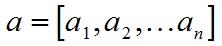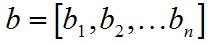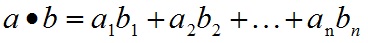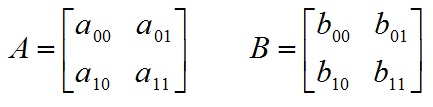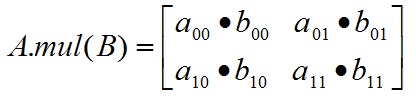SVM介绍
def L(x,y,w):
scores = W.dot(x)
margins = np.maximum(0,scores - scores[y] + 1)
margins[y] = 0
loss_i = np.sum(margins)
return loss_i
linear_svm.py
import numpy as np
from random import shuffle
def svm_loss_naive(W, X, y, reg):
"""
Structured SVM loss function, naive implementation (with loops).
Inputs have dimension D, there are C classes, and we operate on minibatches
of N examples.
Inputs:
- W: A numpy array of shape (D, C) containing weights.
- X: A numpy array of shape (N, D) containing a minibatch of data.
- y: A numpy array of shape (N,) containing training labels; y[i] = c means
that X[i] has label c, where 0 <= c < C.
- reg: (float) regularization strength 正则化强度
Returns a tuple of:
- loss as single float
- gradient with respect to weights W; an array of same shape as W
"""
dW = np.zeros(W.shape) # initialize the gradient as zero
# dw (3073,10) 3073=3072+1 因为预处理(添加一列1作为偏置维度,这样我们在优化时候只要考虑一个权重矩阵W就可以啦.)
# compute the loss and the gradient
num_classes = W.shape[1] #10
num_train = X.shape[0] #X (500,3073)
loss = 0.0
for i in range(num_train):
scores = X[i].dot(W)
# scores (10,1) 代表对于每一类的分数
correct_class_score = scores[y[i]]
#y[i]=c 表示 x[i]的标签为c,其中 0 <= c <= C,correct_class_score即正确标签那一类的分数
for j in range(num_classes):
if j == y[i]: #跳过同类的那一个
continue
margin = scores[j] - correct_class_score + 1 # note delta = 1
if margin > 0:
loss += margin
dW[:, y[i]] += -X[i, :] # 根据公式:∇Wyi Li = - xiT(∑j≠yi1(xiWj - xiWyi +1>0)) + 2λWyi
dW[:, j] += X[i, :] # 根据公式: ∇Wj Li = xiT 1(xiWj - xiWyi +1>0) + 2λWj , (j≠yi)
# Right now the loss is a sum over all training examples, but we want it
# to be an average instead so we divide by num_train.
loss /= num_train
dW /= num_train
# Add regularization to the loss.
loss += reg * np.sum(W * W)
dW += reg * W
#############################################################################
# TODO: #
# Compute the gradient of the loss function and store it dW. #
# Rather that first computing the loss and then computing the derivative, #
# it may be simpler to compute the derivative at the same time that the #
# loss is being computed. As a result you may need to modify some of the #
# code above to compute the gradient. #
#############################################################################
return loss, dW
def svm_loss_vectorized(W, X, y, reg):
"""
Structured SVM loss function, vectorized implementation.
Inputs and outputs are the same as svm_loss_naive.
"""
loss = 0.0
dW = np.zeros(W.shape) # initialize the gradient as zero
#############################################################################
# TODO: #
# Implement a vectorized version of the structured SVM loss, storing the #
# result in loss. #
#############################################################################
scores = X.dot(W)
#scores(500,10)即(N,C),存储每个类的分数
num_classes = W.shape[1]
num_train = X.shape[0] #num_train = N
scores_correct = scores[np.arange(num_train),y]
#scores_correct(1,N) y(N,1) 即对于每个训练样本,找到正确标签那一类的分数 np.arange(num_train)和y分别作为scores的系数
scores_correct = np.reshape(scores_correct,(num_train,-1))
#scores_correct(N,1)
margins = scores - scores_correct + 1
# scores(N,C) scores_correct(N,1) 减法的方法是scores[i]中每一维都减去scores_correct[i]
# margins(N,C)
margins = np.maximum(0,margins)
#对每个元素取max(0,x)
margins[np.arange(num_train),y] = 0
# 跳过同类的那一个
loss += np.sum(margins) / num_train #要除以训练集个数
loss += 0.5 * reg * np.sum(W * W) #正则项
#############################################################################
# END OF YOUR CODE #
#############################################################################
#############################################################################
# TODO: #
# Implement a vectorized version of the gradient for the structured SVM #
# loss, storing the result in dW. #
# #
# Hint: Instead of computing the gradient from scratch, it may be easier #
# to reuse some of the intermediate values that you used to compute the #
# loss. #
#############################################################################
margins[margins > 0] = 1
#大于0的采用求导数
row_sum = np.sum(margins,axis = 1)
#row_sum(1,N) margins中每一行的和
margins[np.arange(num_train),y] = -row_sum
#对于正确标签那一类的梯度计算不同于其它类
dW += np.dot(X.T,margins)/num_train + reg*W
#############################################################################
# END OF YOUR CODE #
#############################################################################
return loss, dW
linear_classifier.py
使用随机梯度下降来训练
from __future__ import print_function
import numpy as np
from cs231n.classifiers.linear_svm import *
from cs231n.classifiers.softmax import *
class LinearClassifier(object):
def __init__(self):
self.W = None
def train(self, X, y, learning_rate=1e-3, reg=1e-5, num_iters=100,
batch_size=200, verbose=False):
#verbose 若为真,优化时打印过程。
"""
Train this linear classifier using stochastic gradient descent.
Inputs:
- X: A numpy array of shape (N, D) containing training data; there are N
training samples each of dimension D.
- y: A numpy array of shape (N,) containing training labels; y[i] = c
means that X[i] has label 0 <= c < C for C classes.
- learning_rate: (float) learning rate for optimization.
- reg: (float) regularization strength.
- num_iters: (integer) number of steps to take when optimizing
- batch_size: (integer) number of training examples to use at each step.
- verbose: (boolean) If true, print progress during optimization.
Outputs:
A list containing the value of the loss function at each training iteration.
"""
num_train, dim = X.shape
num_classes = np.max(y) + 1 # assume y takes values 0...K-1 where K is number of classes
if self.W is None:
# lazily initialize W
self.W = 0.001 * np.random.randn(dim, num_classes)
# Run stochastic gradient descent to optimize W
loss_history = []
for it in range(num_iters):
X_batch = None
y_batch = None
#########################################################################
# TODO: #
# Sample batch_size elements from the training data and their #
# corresponding labels to use in this round of gradient descent. #
# Store the data in X_batch and their corresponding labels in #
# y_batch; after sampling X_batch should have shape (dim, batch_size) #
# and y_batch should have shape (batch_size,) #
# #
# Hint: Use np.random.choice to generate indices. Sampling with #
# replacement is faster than sampling without replacement. #
#########################################################################
batch_inx = np.random.choice(num_train,batch_size)
X_batch = X[batch_inx,:]
y_batch = y[batch_inx]
#采样
#########################################################################
# END OF YOUR CODE #
#########################################################################
# evaluate loss and gradient
loss, grad = self.loss(X_batch, y_batch, reg)
loss_history.append(loss)
# perform parameter update
#########################################################################
# TODO: #
# Update the weights using the gradient and the learning rate. #
#########################################################################
self.W = self.W - learning_rate * grad
#########################################################################
# END OF YOUR CODE #
#########################################################################
if verbose and it % 100 == 0:
print('iteration %d / %d: loss %f' % (it, num_iters, loss))
return loss_history
def predict(self, X):
"""
Use the trained weights of this linear classifier to predict labels for
data points.
Inputs:
- X: A numpy array of shape (N, D) containing training data; there are N
training samples each of dimension D.
Returns:
- y_pred: Predicted labels for the data in X. y_pred is a 1-dimensional
array of length N, and each element is an integer giving the predicted
class.
"""
y_pred = np.zeros(X.shape[0])
###########################################################################
# TODO: #
# Implement this method. Store the predicted labels in y_pred. #
###########################################################################
y_pred = np.argmax(np.dot(X,self.W),axis = 1)
# X(N,D) W(D,C) y_pred(N,1)
###########################################################################
# END OF YOUR CODE #
###########################################################################
return y_pred
def loss(self, X_batch, y_batch, reg):
"""
Compute the loss function and its derivative.
Subclasses will override this.
Inputs:
- X_batch: A numpy array of shape (N, D) containing a minibatch of N
data points; each point has dimension D.
- y_batch: A numpy array of shape (N,) containing labels for the minibatch.
- reg: (float) regularization strength.
Returns: A tuple containing:
- loss as a single float
- gradient with respect to self.W; an array of the same shape as W
"""
pass
class LinearSVM(LinearClassifier):
""" A subclass that uses the Multiclass SVM loss function """
def loss(self, X_batch, y_batch, reg):
return svm_loss_vectorized(self.W, X_batch, y_batch, reg)
class Softmax(LinearClassifier):
""" A subclass that uses the Softmax + Cross-entropy loss function """
def loss(self, X_batch, y_batch, reg):
return softmax_loss_vectorized(self.W, X_batch, y_batch, reg)
cross-validation部分:
# Use the validation set to tune hyperparameters (regularization strength and
# learning rate). You should experiment with different ranges for the learning
# rates and regularization strengths; if you are careful you should be able to
# get a classification accuracy of about 0.4 on the validation set.
learning_rates = [1e-7, 5e-5]
regularization_strengths = [2.5e4, 5e4]
# results is dictionary mapping tuples of the form
# (learning_rate, regularization_strength) to tuples of the form
# (training_accuracy, validation_accuracy). The accuracy is simply the fraction
# of data points that are correctly classified.
results = {}
best_val = -1 # The highest validation accuracy that we have seen so far.
best_svm = None # The LinearSVM object that achieved the highest validation rate.
################################################################################
# TODO: #
# Write code that chooses the best hyperparameters by tuning on the validation #
# set. For each combination of hyperparameters, train a linear SVM on the #
# training set, compute its accuracy on the training and validation sets, and #
# store these numbers in the results dictionary. In addition, store the best #
# validation accuracy in best_val and the LinearSVM object that achieves this #
# accuracy in best_svm. #
# #
# Hint: You should use a small value for num_iters as you develop your #
# validation code so that the SVMs don't take much time to train; once you are #
# confident that your validation code works, you should rerun the validation #
# code with a larger value for num_iters. #
################################################################################
for rate in learning_rates:
for regular in regularization_strengths:
svm = LinearSVM()
svm.train(X_train,y_train,learning_rate = rate,reg = regular,num_iters = 1000)
y_train_pred = svm.predict(X_train)
accuracy_train = np.mean(y_train == y_train_pred)
y_val_pred = svm.predict(X_val)
accuracy_val = np.mean(y_val == y_val_pred)
results[(rate,regular)] = (accuracy_train,accuracy_val)
if best_val < accuracy_val:
best_val = accuracy_val
best_svm = svm
################################################################################
# END OF YOUR CODE #
################################################################################
# Print out results.
for lr, reg in sorted(results):
train_accuracy, val_accuracy = results[(lr, reg)]
print('lr %e reg %e train accuracy: %f val accuracy: %f' % (
lr, reg, train_accuracy, val_accuracy))
print('best validation accuracy achieved during cross-validation: %f' % best_val)
Note:
-
- dot mul
- A*B 矩阵乘法,返回矩阵
- A.dot(B) 內积,返回一个标量
- A.mul(B) 要求A和B的形状一致,对应元素的乘法。
参考:https://blog.csdn.net/dcrmg/article/details/52404580
- np.arrange
返回值: np.arange()函数返回一个有终点和起点的固定步长的排列,如[1,2,3,4,5],起点是1,终点是5,步长为1。
参数个数情况: np.arange()函数分为一个参数,两个参数,三个参数三种情况
1)一个参数时,参数值为终点,起点取默认值0,步长取默认值1。
2)两个参数时,第一个参数为起点,第二个参数为终点,步长取默认值1。
3)三个参数时,第一个参数为起点,第二个参数为终点,第三个参数为步长。其中步长支持小数。
#一个参数 默认起点0,步长为1 输出:[0 1 2]
a = np.arange(3)
#两个参数 默认步长为1 输出[3 4 5 6 7 8]
a = np.arange(3,9)
#三个参数 起点为0,终点为4,步长为0.1 输出[ 0. 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1. 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 2. 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9]
a = np.arange(0, 3, 0.1)
参考:https://blog.csdn.net/u011649885/article/details/76851291
- np.random.choice
import numpy as np
# 参数意思分别 是从a 中以概率P,随机选择3个, p没有指定的时候相当于是一致的分布
a1 = np.random.choice(a=5, size=3, replace=False, p=None)
print(a1)
# 非一致的分布,会以多少的概率提出来
a2 = np.random.choice(a=5, size=3, replace=False, p=[0.2, 0.1, 0.3, 0.4, 0.0])
print(a2)
# replacement 代表的意思是抽样之后还放不放回去,如果是False的话,那么出来的三个数都不一样,如果是
True的话, 有可能会出现重复的,因为前面的抽的放回去了。