二分类:SVMs,logistic regression,decision trees,random forests,gradient-boosted trees,naive Bayes
多分类: logistic regression,decision trees,random forests, naive Bayes
归回: linear least regression, decision tress,random forests,gradient-boosted trees, isotonic regression。
一。Linear models
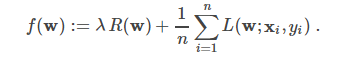

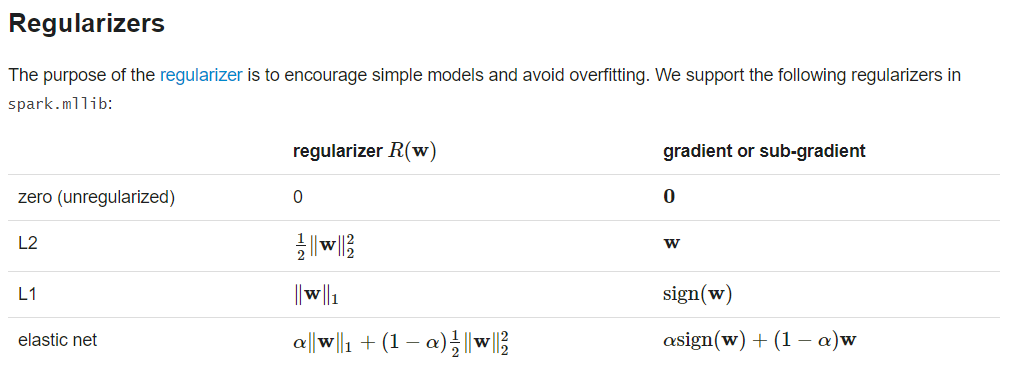
classification (SVMs, logistic regression)

1 package ML.ClassificationAndRegression; 2 3 import org.apache.spark.SparkConf; 4 import org.apache.spark.api.java.JavaRDD; 5 import org.apache.spark.api.java.JavaSparkContext; 6 import org.apache.spark.api.java.function.Function; 7 import org.apache.spark.mllib.classification.SVMModel; 8 import org.apache.spark.mllib.classification.SVMWithSGD; 9 import org.apache.spark.mllib.evaluation.BinaryClassificationMetrics; 10 import org.apache.spark.mllib.optimization.L1Updater; 11 import org.apache.spark.mllib.regression.LabeledPoint; 12 import org.apache.spark.mllib.util.MLUtils; 13 import org.apache.spark.rdd.RDD; 14 import scala.Tuple2; 15 16 /** 17 * TODO 18 * 19 * @ClassName: SVMClassifier 20 * @author: DingH 21 * @since: 2019/4/9 10:28 22 */ 23 public class SVMClassifier { 24 public static void main(String[] args) { 25 SparkConf conf = new SparkConf().setAppName("SVM Classifier Example").setMaster("local"); 26 JavaSparkContext jsc = new JavaSparkContext(conf); 27 String path = "D:\IdeaProjects\SimpleApp\src\main\resources\data\mllib\sample_libsvm_data.txt"; 28 JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), path).toJavaRDD(); 29 30 // Split initial RDD into two... [60% training data, 40% testing data]. 31 JavaRDD<LabeledPoint> train = data.sample(false, 0.6, 11L); 32 train.cache(); 33 final JavaRDD<LabeledPoint> test = data.subtract(train); 34 35 //Run training algorithm to build the model 36 int numsIterations = 100; 37 SVMWithSGD svm = new SVMWithSGD(); 38 svm.optimizer().setNumIterations(200).setRegParam(0.01).setUpdater(new L1Updater()); 39 final SVMModel model1 = svm.run(train.rdd()); 40 // final SVMModel model1 = SVMWithSGD.train(train.rdd(), numsIterations); 41 42 model1.clearThreshold(); 43 44 JavaRDD<Tuple2<Object, Object>> scoraAndLables = test.map(new Function<LabeledPoint, Tuple2<Object, Object>>() { 45 public Tuple2<Object, Object> call(LabeledPoint p) throws Exception { 46 double predict = model1.predict(p.features()); 47 return new Tuple2<Object, Object>(predict, p.label()); 48 } 49 }); 50 51 BinaryClassificationMetrics metrics = new BinaryClassificationMetrics(scoraAndLables.rdd()); 52 53 double areaUnderROC = metrics.areaUnderROC(); 54 55 System.out.println("Area under ROC = " + areaUnderROC); 56 57 model1.save(jsc.sc(),"D:\IdeaProjects\SimpleApp\src\main\java\MLModel"); 58 SVMModel model = SVMModel.load(jsc.sc(), "D:\IdeaProjects\SimpleApp\src\main\java\MLModel"); 59 60 } 61 }

1 package ML.ClassificationAndRegression; 2 3 import org.apache.spark.SparkConf; 4 import org.apache.spark.api.java.JavaRDD; 5 import org.apache.spark.api.java.JavaSparkContext; 6 import org.apache.spark.api.java.function.Function; 7 import org.apache.spark.ml.classification.MultiClassSummarizer; 8 import org.apache.spark.mllib.classification.LogisticRegressionModel; 9 import org.apache.spark.mllib.classification.LogisticRegressionWithLBFGS; 10 import org.apache.spark.mllib.classification.LogisticRegressionWithSGD; 11 import org.apache.spark.mllib.evaluation.MulticlassMetrics; 12 import org.apache.spark.mllib.regression.LabeledPoint; 13 import org.apache.spark.mllib.util.MLUtils; 14 import scala.Tuple2; 15 16 /** 17 * TODO 18 * 19 * @ClassName: LogistiRegression 20 * @author: DingH 21 * @since: 2019/4/9 11:08 22 */ 23 public class LogistiRegression { 24 public static void main(String[] args) { 25 SparkConf conf = new SparkConf().setMaster("local").setAppName("LogisticRegression"); 26 JavaSparkContext jsc = new JavaSparkContext(conf); 27 String path = "D:\IdeaProjects\SimpleApp\src\main\resources\data\mllib\sample_libsvm_data.txt"; 28 JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), path).toJavaRDD(); 29 30 JavaRDD<LabeledPoint>[] split = data.randomSplit(new double[]{0.6, 0.4}, 11L); 31 JavaRDD<LabeledPoint> training = split[0].cache(); 32 final JavaRDD<LabeledPoint> test = split[1]; 33 34 final LogisticRegressionModel model = new LogisticRegressionWithLBFGS().setNumClasses(10).run(training.rdd()); 35 JavaRDD<Tuple2<Object, Object>> predictionAndLabels = test.map(new Function<LabeledPoint, Tuple2<Object, Object>>() { 36 public Tuple2<Object, Object> call(LabeledPoint labeledPoint) throws Exception { 37 double predict = model.predict(labeledPoint.features()); 38 return new Tuple2<Object, Object>(predict, labeledPoint.label()); 39 } 40 }); 41 42 MulticlassMetrics metrics = new MulticlassMetrics(predictionAndLabels.rdd()); 43 44 double precision = metrics.precision(); 45 System.out.println("Precision = " + precision); 46 47 // Save and load model 48 // model.save(jsc.sc(), "myModelPath"); 49 // LogisticRegressionModel sameModel = LogisticRegressionModel.load(jsc.sc(), "myModelPath"); 50 } 51 }
linear regression (least squares, Lasso, ridge)

1 package ML.ClassificationAndRegression; 2 3 import org.apache.hadoop.yarn.webapp.hamlet.Hamlet; 4 import org.apache.spark.SparkConf; 5 import org.apache.spark.api.java.JavaDoubleRDD; 6 import org.apache.spark.api.java.JavaRDD; 7 import org.apache.spark.api.java.JavaSparkContext; 8 import org.apache.spark.api.java.function.Function; 9 import org.apache.spark.mllib.linalg.Vectors; 10 import org.apache.spark.mllib.regression.LabeledPoint; 11 import org.apache.spark.mllib.regression.LinearRegressionModel; 12 import org.apache.spark.mllib.regression.LinearRegressionWithSGD; 13 import org.apache.spark.mllib.util.MLUtils; 14 import scala.Tuple2; 15 16 /** 17 * TODO 18 * 19 * @ClassName: Regression 20 * @author: DingH 21 * @since: 2019/4/9 11:21 22 */ 23 public class Regression { 24 public static void main(String[] args) { 25 SparkConf conf = new SparkConf().setAppName("Regression").setMaster("local"); 26 JavaSparkContext jsc = new JavaSparkContext(conf); 27 String path = "D:\IdeaProjects\SimpleApp\src\main\resources\data\mllib\ridge-data\lpsa.data"; 28 JavaRDD<String> data = jsc.textFile(path); 29 30 JavaRDD<LabeledPoint> parsedData = data.map(new Function<String, LabeledPoint>() { 31 public LabeledPoint call(String line) throws Exception { 32 String[] split = line.split(","); 33 String[] features = split[1].split(" "); 34 double[] v = new double[features.length]; 35 for (int i = 0; i < features.length - 1; i++) { 36 v[i] = Double.parseDouble(features[i]); 37 } 38 39 return new LabeledPoint(Double.parseDouble(split[0]), Vectors.dense(v)); 40 } 41 }).cache(); 42 43 final LinearRegressionModel model = LinearRegressionWithSGD.train(parsedData.rdd(), 100); 44 45 JavaRDD<Tuple2<Double, Double>> valuesAndLabels = parsedData.map(new Function<LabeledPoint, Tuple2<Double, Double>>() { 46 public Tuple2<Double, Double> call(LabeledPoint labeledPoint) throws Exception { 47 double predict = model.predict(labeledPoint.features()); 48 return new Tuple2<Double, Double>(predict, labeledPoint.label()); 49 } 50 }); 51 52 Double MSE = new JavaDoubleRDD(valuesAndLabels.map( 53 new Function<Tuple2<Double, Double>, Object>() { 54 public Object call(Tuple2<Double, Double> dat) throws Exception { 55 return Math.pow(dat._1 - dat._2, 2.0); 56 } 57 } 58 ).rdd()).mean(); 59 System.out.println("training Mean Squared Error = " + MSE); 60 61 // Save and load model 62 // model.save(jsc.sc(), "myModelPath"); 63 // LinearRegressionModel sameModel = LinearRegressionModel.load(jsc.sc(), "myModelPath"); 64 } 65 }
二。Decision Trees.
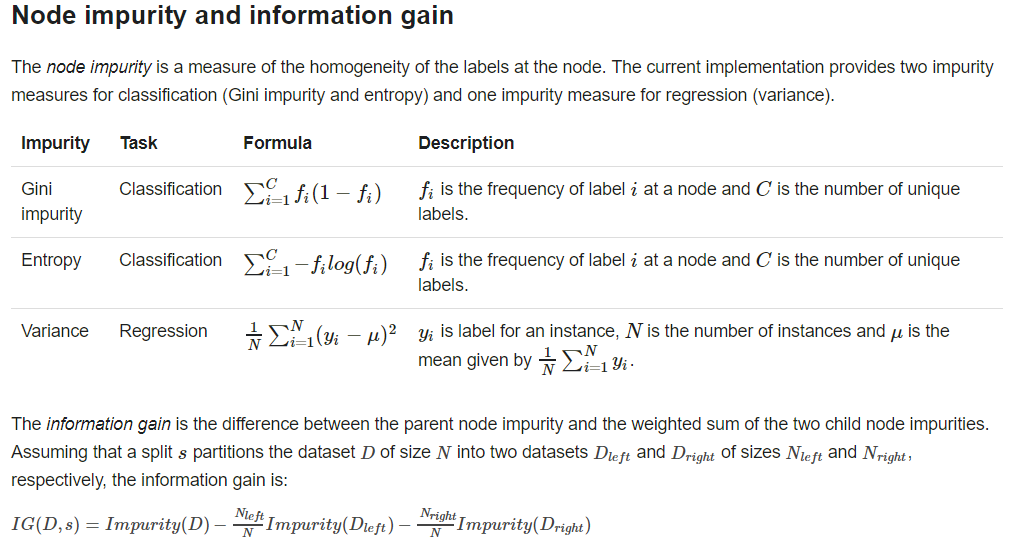
problem specification parameters: algo, numClasses, categoricalFeaturesInfo
stopping criteria : maxDepth, minInfoGain, minInstancePerNode
tunnable parameters: maxBins, impurity,

package ML.DT; import org.apache.spark.SparkConf; import org.apache.spark.api.java.JavaPairRDD; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.JavaSparkContext; import org.apache.spark.api.java.function.Function; import org.apache.spark.api.java.function.PairFunction; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.tree.DecisionTree; import org.apache.spark.mllib.tree.model.DecisionTreeModel; import org.apache.spark.mllib.util.MLUtils; import scala.Tuple2; import java.util.HashMap; /** * TODO * * @ClassName: classification * @author: DingH * @since: 2019/4/9 16:11 */ public class classification { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("DTclassification").setMaster("local"); JavaSparkContext jsc = new JavaSparkContext(conf); String path = "D:\IdeaProjects\SimpleApp\src\main\resources\data\mllib\sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), path).toJavaRDD(); JavaRDD<LabeledPoint>[] split = data.randomSplit(new double[]{0.7, 0.3}, 11L); JavaRDD<LabeledPoint> trainningData = split[0]; JavaRDD<LabeledPoint> test = split[1]; int numsClasses = 2; HashMap<Integer, Integer> categoricalFeaturesInfo = new HashMap<Integer, Integer>(); String impurity = "gini"; int maxDepth = 1; int maxbins = 32; final DecisionTreeModel model = DecisionTree.trainClassifier(trainningData, numsClasses,categoricalFeaturesInfo, impurity, maxDepth,maxbins); JavaPairRDD<Double, Double> predictionAndLable = test.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { public Tuple2<Double, Double> call(LabeledPoint labeledPoint) throws Exception { return new Tuple2<Double, Double>(model.predict(labeledPoint.features()), labeledPoint.label()); } }); double testErr = predictionAndLable.filter(new Function<Tuple2<Double, Double>, Boolean>() { public Boolean call(Tuple2<Double, Double> doubleDoubleTuple2) throws Exception { return !doubleDoubleTuple2._1().equals(doubleDoubleTuple2._2()); } }).count() * 1.0 / test.count(); System.out.println("Test Error: " + testErr); System.out.println("Learned classification tree model: " + model.toDebugString()); // Save and load model // model.save(jsc.sc(), "target/tmp/myDecisionTreeClassificationModel"); // DecisionTreeModel sameModel = DecisionTreeModel.load(jsc.sc(), "target/tmp/myDecisionTreeClassificationModel"); } }

package ML.DT; import org.apache.spark.SparkConf; import org.apache.spark.api.java.JavaPairRDD; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.JavaSparkContext; import org.apache.spark.api.java.function.Function; import org.apache.spark.api.java.function.Function2; import org.apache.spark.api.java.function.PairFunction; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.tree.DecisionTree; import org.apache.spark.mllib.tree.model.DecisionTreeModel; import org.apache.spark.mllib.util.MLUtils; import scala.Tuple2; import java.util.HashMap; import java.util.Map; /** * TODO * * @ClassName: Regression * @author: DingH * @since: 2019/4/9 16:33 */ public class Regression { public static void main(String[] args) { SparkConf sparkConf = new SparkConf().setAppName("JavaDecisionTreeRegressionExample").setMaster("local"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "D:\IdeaProjects\SimpleApp\src\main\resources\data\mllib\sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Set parameters. // Empty categoricalFeaturesInfo indicates all features are continuous. Map<Integer, Integer> categoricalFeaturesInfo = new HashMap<Integer, Integer>(); String impurity = "variance"; Integer maxDepth = 5; Integer maxBins = 32; // Train a DecisionTree model. final DecisionTreeModel model = DecisionTree.trainRegressor(trainingData, categoricalFeaturesInfo, impurity, maxDepth, maxBins); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<Double, Double>(model.predict(p.features()), p.label()); } }); Double testMSE = predictionAndLabel.map(new Function<Tuple2<Double, Double>, Double>() { public Double call(Tuple2<Double, Double> pl) { Double diff = pl._1() - pl._2(); return diff * diff; } }).reduce(new Function2<Double, Double, Double>() { public Double call(Double a, Double b) { return a + b; } }) / data.count(); System.out.println("Test Mean Squared Error: " + testMSE); System.out.println("Learned regression tree model: " + model.toDebugString()); // Save and load model // model.save(jsc.sc(), "target/tmp/myDecisionTreeRegressionModel"); // DecisionTreeModel sameModel = DecisionTreeModel.load(jsc.sc(), "target/tmp/myDecisionTreeRegressionModel"); } }
三。Random Forests
样本随机,特征随机
featureSubsetStrategy - Number of features to consider for splits at each node. Supported: "auto", "all", "sqrt", "log2", "onethird". If "auto" is set, this parameter is set based on numTrees: if numTrees == 1, set to "all"; if numTrees > 1 (forest) set to "sqrt".

package ML.RF; import org.apache.spark.SparkConf; import org.apache.spark.api.java.JavaPairRDD; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.JavaSparkContext; import org.apache.spark.api.java.function.Function; import org.apache.spark.api.java.function.PairFunction; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.tree.RandomForest; import org.apache.spark.mllib.tree.model.RandomForestModel; import org.apache.spark.mllib.util.MLUtils; import scala.Tuple2; import java.util.HashMap; /** * TODO * * @ClassName: classification * @author: DingH * @since: 2019/4/9 16:58 */ public class classification { public static void main(String[] args) { SparkConf sparkConf = new SparkConf().setMaster("local").setAppName("JavaRandomForestClassificationExample"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "D:\IdeaProjects\SimpleApp\src\main\resources\data\mllib\sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Train a RandomForest model. // Empty categoricalFeaturesInfo indicates all features are continuous. Integer numClasses = 2; HashMap<Integer, Integer> categoricalFeaturesInfo = new HashMap<Integer, Integer>(); Integer numTrees = 3; // Use more in practice. String featureSubsetStrategy = "auto"; // Let the algorithm choose. String impurity = "gini"; Integer maxDepth = 5; Integer maxBins = 32; Integer seed = 12345; final RandomForestModel model = RandomForest.trainClassifier(trainingData, numClasses,categoricalFeaturesInfo, numTrees, featureSubsetStrategy, impurity, maxDepth, maxBins, seed); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<Double, Double>(model.predict(p.features()), p.label()); } }); Double testErr = 1.0 * predictionAndLabel.filter(new Function<Tuple2<Double, Double>, Boolean>() { public Boolean call(Tuple2<Double, Double> pl) { return !pl._1().equals(pl._2()); } }).count() / testData.count(); System.out.println("Test Error: " + testErr); System.out.println("Learned classification forest model: " + model.toDebugString()); // Save and load model // model.save(jsc.sc(), "target/tmp/myRandomForestClassificationModel"); // RandomForestModel sameModel = RandomForestModel.load(jsc.sc(),"target/tmp/myRandomForestClassificationModel"); } }

package ML.RF; import org.apache.spark.SparkConf; import org.apache.spark.api.java.JavaPairRDD; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.JavaSparkContext; import org.apache.spark.api.java.function.Function; import org.apache.spark.api.java.function.Function2; import org.apache.spark.api.java.function.PairFunction; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.tree.RandomForest; import org.apache.spark.mllib.tree.model.RandomForestModel; import org.apache.spark.mllib.util.MLUtils; import scala.Tuple2; import java.util.HashMap; import java.util.Map; /** * TODO * * @ClassName: regression * @author: DingH * @since: 2019/4/9 17:50 */ public class regression { public static void main(String[] args) { SparkConf sparkConf = new SparkConf().setMaster("local").setAppName("JavaRandomForestRegressionExample"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "D:\IdeaProjects\SimpleApp\src\main\resources\data\mllib\sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Set parameters. // Empty categoricalFeaturesInfo indicates all features are continuous. Map<Integer, Integer> categoricalFeaturesInfo = new HashMap<Integer, Integer>(); Integer numTrees = 3; // Use more in practice. String featureSubsetStrategy = "auto"; // Let the algorithm choose. String impurity = "variance"; Integer maxDepth = 4; Integer maxBins = 32; Integer seed = 12345; // Train a RandomForest model. final RandomForestModel model = RandomForest.trainRegressor(trainingData,categoricalFeaturesInfo, numTrees, featureSubsetStrategy, impurity, maxDepth, maxBins, seed); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<Double, Double>(model.predict(p.features()), p.label()); } }); Double testMSE = predictionAndLabel.map(new Function<Tuple2<Double, Double>, Double>() { public Double call(Tuple2<Double, Double> pl) { Double diff = pl._1() - pl._2(); return diff * diff; } }).reduce(new Function2<Double, Double, Double>() { public Double call(Double a, Double b) { return a + b; } }) / testData.count(); System.out.println("Test Mean Squared Error: " + testMSE); System.out.println("Learned regression forest model: " + model.toDebugString()); // Save and load model model.save(jsc.sc(), "target/tmp/myRandomForestRegressionModel"); RandomForestModel sameModel = RandomForestModel.load(jsc.sc(), "target/tmp/myRandomForestRegressionModel"); } }
四。Gradient-Boosted Trees

Usage tips: loss, numIterations, learningRate, algo
BoostingStrategy.validationTol

package ML.GradientBoostedTrees; import org.apache.spark.SparkConf; import org.apache.spark.api.java.JavaPairRDD; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.JavaSparkContext; import org.apache.spark.api.java.function.Function; import org.apache.spark.api.java.function.PairFunction; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.tree.GradientBoostedTrees; import org.apache.spark.mllib.tree.configuration.BoostingStrategy; import org.apache.spark.mllib.tree.model.GradientBoostedTreesModel; import org.apache.spark.mllib.util.MLUtils; import scala.Tuple2; import java.util.HashMap; import java.util.Map; /** * TODO * * @ClassName: classification * @author: DingH * @since: 2019/4/9 17:56 */ public class classification { public static void main(String[] args) { SparkConf sparkConf = new SparkConf().setMaster("local").setAppName("JavaGradientBoostedTreesClassificationExample"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "D:\IdeaProjects\SimpleApp\src\main\resources\data\mllib\sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Train a GradientBoostedTrees model. // The defaultParams for Classification use LogLoss by default. BoostingStrategy boostingStrategy = BoostingStrategy.defaultParams("Classification"); boostingStrategy.setNumIterations(3); // Note: Use more iterations in practice. boostingStrategy.getTreeStrategy().setNumClasses(2); boostingStrategy.getTreeStrategy().setMaxDepth(5); // Empty categoricalFeaturesInfo indicates all features are continuous. Map<Integer, Integer> categoricalFeaturesInfo = new HashMap<Integer, Integer>(); boostingStrategy.treeStrategy().setCategoricalFeaturesInfo(categoricalFeaturesInfo); final GradientBoostedTreesModel model = GradientBoostedTrees.train(trainingData, boostingStrategy); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<Double, Double>(model.predict(p.features()), p.label()); } }); Double testErr = 1.0 * predictionAndLabel.filter(new Function<Tuple2<Double, Double>, Boolean>() { public Boolean call(Tuple2<Double, Double> pl) { return !pl._1().equals(pl._2()); } }).count() / testData.count(); System.out.println("Test Error: " + testErr); System.out.println("Learned classification GBT model: " + model.toDebugString()); // Save and load model // model.save(jsc.sc(), "target/tmp/myGradientBoostingClassificationModel"); // GradientBoostedTreesModel sameModel = GradientBoostedTreesModel.load(jsc.sc(), // "target/tmp/myGradientBoostingClassificationModel"); } }
 regression
regression五。naive Bayes
model type : "multinomial","bernouli"

1 package ML.ClassificationAndRegression.NaiveBayes; 2 3 import org.apache.spark.SparkConf; 4 import org.apache.spark.api.java.JavaRDD; 5 import org.apache.spark.api.java.JavaSparkContext; 6 import org.apache.spark.api.java.function.Function; 7 import org.apache.spark.mllib.classification.NaiveBayes; 8 import org.apache.spark.mllib.classification.NaiveBayesModel; 9 import org.apache.spark.mllib.regression.LabeledPoint; 10 import org.apache.spark.mllib.util.MLUtils; 11 import scala.Tuple2; 12 13 /** 14 * TODO 15 * 16 * @ClassName: example 17 * @author: DingH 18 * @since: 2019/4/10 10:04 19 */ 20 public class example { 21 public static void main(String[] args) { 22 SparkConf conf = new SparkConf().setMaster("local").setAppName("naiveBayesExample"); 23 JavaSparkContext jsc = new JavaSparkContext(conf); 24 25 String path = "D:\IdeaProjects\SimpleApp\src\main\resources\data\mllib\sample_libsvm_data.txt"; 26 JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), path).toJavaRDD(); 27 28 JavaRDD<LabeledPoint>[] split = data.randomSplit(new double[]{0.6, 0.4}, 12345L); 29 JavaRDD<LabeledPoint> train = split[0]; 30 JavaRDD<LabeledPoint> test = split[1]; 31 32 final NaiveBayesModel model = NaiveBayes.train(train.rdd(), 1.0, "multinomial"); 33 34 JavaRDD<Tuple2<Double, Double>> predictionAndLabel = test.map(new Function<LabeledPoint, Tuple2<Double, Double>>() { 35 public Tuple2<Double, Double> call(LabeledPoint labeledPoint) throws Exception { 36 double predict = model.predict(labeledPoint.features()); 37 return new Tuple2<Double, Double>(predict, labeledPoint.label()); 38 } 39 }); 40 41 double accuracy = predictionAndLabel.filter(new Function<Tuple2<Double, Double>, Boolean>() { 42 public Boolean call(Tuple2<Double, Double> doubleDoubleTuple2) throws Exception { 43 return doubleDoubleTuple2._1().equals(doubleDoubleTuple2._2()); 44 } 45 }).count() / (double) test.count(); 46 47 System.out.println("acucuracy is : " + accuracy); 48 49 NaiveBayesModel model1 = NaiveBayesModel.load(jsc.sc(), ""); 50 51 52 } 53 }
六。isotonic regression
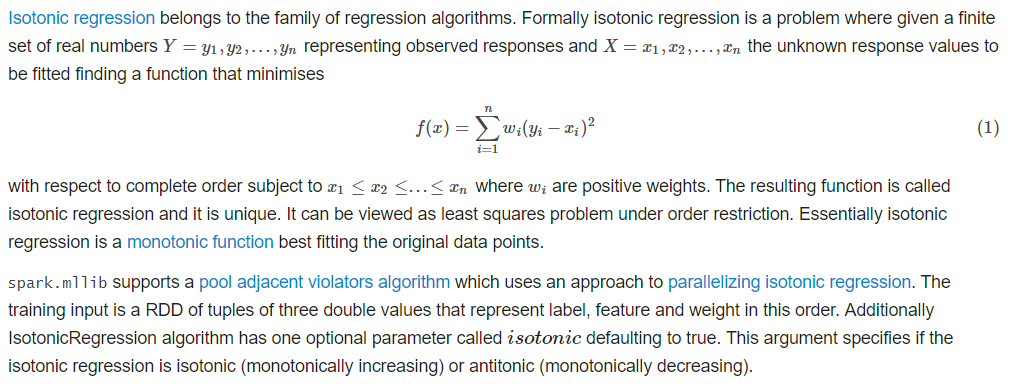

1 package ML.ClassificationAndRegression.IsotonicRegression; 2 3 import org.apache.spark.SparkConf; 4 import org.apache.spark.api.java.JavaDoubleRDD; 5 import org.apache.spark.api.java.JavaRDD; 6 import org.apache.spark.api.java.JavaSparkContext; 7 import org.apache.spark.api.java.function.Function; 8 import org.apache.spark.mllib.regression.IsotonicRegression; 9 import org.apache.spark.mllib.regression.IsotonicRegressionModel; 10 import scala.Tuple2; 11 import scala.Tuple3; 12 13 /** 14 * TODO 15 * 16 * @ClassName: example 17 * @author: DingH 18 * @since: 2019/4/10 10:31 19 */ 20 public class example { 21 public static void main(String[] args) { 22 SparkConf conf = new SparkConf().setMaster("local").setAppName("isotonicRegressionExample"); 23 JavaSparkContext jsc = new JavaSparkContext(conf); 24 25 String path = "D:\IdeaProjects\SimpleApp\src\main\resources\data\mllib\sample_isotonic_regression_data.txt"; 26 JavaRDD<String> data = jsc.textFile(path); 27 28 JavaRDD<Tuple3<Double, Double, Double>> parsedData = data.map(new Function<String, Tuple3<Double, Double, Double>>() { 29 public Tuple3<Double, Double, Double> call(String s) throws Exception { 30 String[] strings = s.split(","); 31 return new Tuple3<Double, Double, Double>(Double.parseDouble(strings[0]), Double.parseDouble(strings[1]), 1.0); 32 } 33 }); 34 35 JavaRDD<Tuple3<Double, Double, Double>>[] split = parsedData.randomSplit(new double[]{0.7, 0.3}, 1234L); 36 JavaRDD<Tuple3<Double, Double, Double>> train = split[0]; 37 JavaRDD<Tuple3<Double, Double, Double>> test = split[1]; 38 39 final IsotonicRegressionModel model = new IsotonicRegression().setIsotonic(true).run(train); 40 41 JavaRDD<Tuple2<Double, Double>> preditionAndLabel = test.map(new Function<Tuple3<Double, Double, Double>, Tuple2<Double, Double>>() { 42 public Tuple2<Double, Double> call(Tuple3<Double, Double, Double> doubleDoubleDoubleTuple3) throws Exception { 43 double predict = model.predict(doubleDoubleDoubleTuple3._1()); 44 return new Tuple2<Double, Double>(predict, doubleDoubleDoubleTuple3._2()); 45 } 46 }); 47 48 Double MSE = new JavaDoubleRDD(preditionAndLabel.map(new Function<Tuple2<Double, Double>, Object>() { 49 public Object call(Tuple2<Double, Double> doubleDoubleTuple2) throws Exception { 50 51 return Math.pow(doubleDoubleTuple2._1() - doubleDoubleTuple2._2(), 2.0); 52 } 53 }).rdd()).mean(); 54 55 System.out.println("Mean Squared Error = " + MSE); 56 } 57 }
