1、数据库

orthodb数据:
odb10v0_levels.tab.gz: NCBI taxonomy nodes where Ortho DB orthologous groups (OGs) are calculated
odb10v0_species.tab.gz: Ortho DB individual organism (aka species) ids based on NCBI taxonomy ids (mostly species level)
odb10v0_level2species.tab.gz: correspondence between level ids and species ids
odb10v0_genes.tab.gz: Ortho DB genes with some info
odb10v0_OGs.tab.gz: Ortho DB orthologous groups
odb10v0_OG2genes.tab.gz: OGs to genes correspondence
odb10v0_OG_xrefs.tab.gz: OG associations with GO, COG and InterPro ids
v9_v10_OGs_map.tab.gz mappings between the previous and current release orthologous group ids
odb10v0_fasta_<root>.tgz tar-ball with one fasta file per taxon id in the given root (bacteria,metazoa,fungi,plants)
2、odb10v0_levels.tab:
1. level NCBI taxonomy id
2. scientific name
3. total non-redundant count of genes in all underneath clustered species(在聚集的物种下面的所有的基因的总非重复计数)
4. total count of OGs built on it
5. total non-redundant count of species underneath

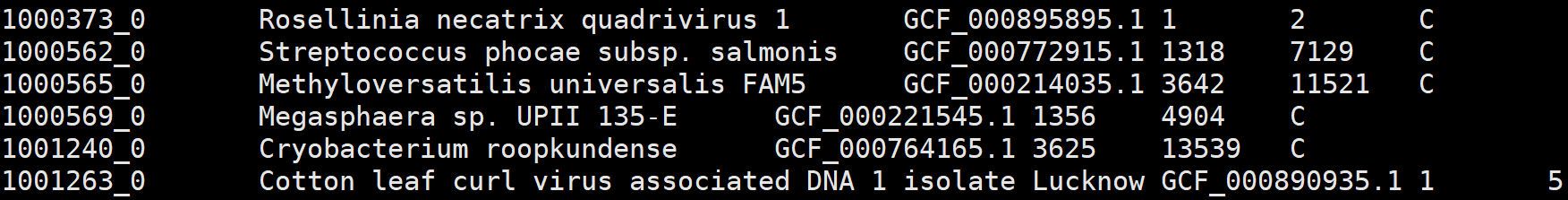
3、odb10v0_species.tab.gz
1. Ortho DB individual organism id, based on NCBI tax id
2. scientific name inherited from the most relevant NCBI tax id
3. genome asssembly id, when available
4. total count of clustered genes in this species
5. total count of the OGs it participates
6. mapping type, clustered(C) or mapped(M)
4、odb10v0_level2species.tab
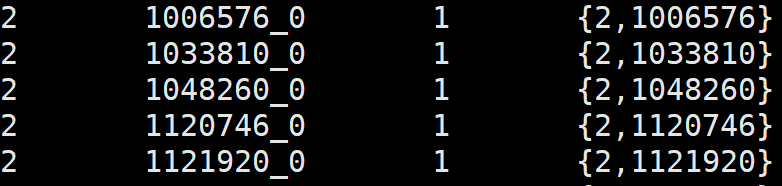
1. top-most level NCBI tax id, one of [2,2157,2759,10239]
2. Ortho DB organism id
3. number of hops between the top-most level id and the NCBI tax id assiciated with the organism
4. ordered list of Ortho DB selected intermediate levels from the top-most level to the bottom one
5、odb10v0_genes.tab
1. Ortho DB unique gene id (not stable between releases)
2. organism tax id
3. protein original sequence id, as downloaded along with the sequence
4. Uniprot id, evaluated by mapping
5. ENSEMBL gene name, evaluated by mapping
6. NCBI gid, evaluated by mapping
7. description, evaluated by mapping
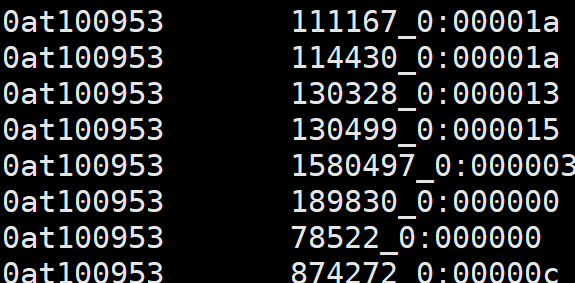
6、odb10v0_OG2genes.tab
1. OG unique id
2. Ortho DB gene id
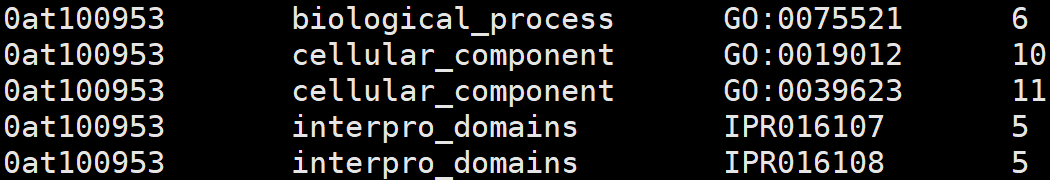
7、odb10v0_OG_xrefs.tab
1. OG unique id
2. external DB or DB section
3. external identifier
4. number of genes in the OG associated with the identifier
参考
https://www.orthodb.org/?page=filelist