tophat输出结果junction.bed
| BED format |
|
BED format provides a flexible way to define the data lines that are displayed in an annotation track. BED lines have three required fields and nine additional optional fields. The number of fields per line must be consistent throughout any single set of data in an annotation track. The order of the optional fields is binding: lower-numbered fields must always be populated if higher-numbered fields are used. If your data set is BED-like, but it is very large and you would like to keep it on your own server, you should use the bigBed data format. The first three required BED fields are:
The 9 additional optional BED fields are:
Example: track name=pairedReads description="Clone Paired Reads" useScore=1 chr22 1000 5000 cloneA 960 + 1000 5000 0 2 567,488, 0,3512 chr22 2000 6000 cloneB 900 - 2000 6000 0 2 433,399, 0,3601 Example: browser position chr7:127471196-127495720 browser hide all track name="ItemRGBDemo" description="Item RGB demonstration" visibility=2 itemRgb="On" chr7 127471196 127472363 Pos1 0 + 127471196 127472363 255,0,0 chr7 127472363 127473530 Pos2 0 + 127472363 127473530 255,0,0 chr7 127473530 127474697 Pos3 0 + 127473530 127474697 255,0,0 chr7 127474697 127475864 Pos4 0 + 127474697 127475864 255,0,0 chr7 127475864 127477031 Neg1 0 - 127475864 127477031 0,0,255 chr7 127477031 127478198 Neg2 0 - 127477031 127478198 0,0,255 chr7 127478198 127479365 Neg3 0 - 127478198 127479365 0,0,255 chr7 127479365 127480532 Pos5 0 + 127479365 127480532 255,0,0 chr7 127480532 127481699 Neg4 0 - 127480532 127481699 0,0,255 Example: browser position chr7:127471196-127495720 browser hide all track name="ColorByStrandDemo" description="Color by strand demonstration" visibility=2 colorByStrand="255,0,0 0,0,255" chr7 127471196 127472363 Pos1 0 + chr7 127472363 127473530 Pos2 0 + chr7 127473530 127474697 Pos3 0 + chr7 127474697 127475864 Pos4 0 + chr7 127475864 127477031 Neg1 0 - chr7 127477031 127478198 Neg2 0 - chr7 127478198 127479365 Neg3 0 - chr7 127479365 127480532 Pos5 0 + chr7 127480532 127481699 Neg4 0 - |
tophat输出结果junction.bed中定义的junction就是中间是intron两边是exon的这种序列结构, JUNC00000001包含两个exon和中间的intron
cat junctions.bed
track name=junctions description="TopHat junctions" test_chromosome 177 400 JUNC00000001 61 + 177 400 255,0,0 2 73,50 0,173 test_chromosome 350 550 JUNC00000002 45 + 350 550 255,0,0 2 50,50 0,150
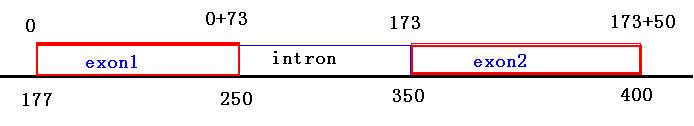
exon1: 177-250(display), 177-249(actual)
intron: 250-350(display), 250-349(actual)
exon2: 350-400(display), 350-399(actual)
chrom(test_chromosome): The name of the chromosome.
这里就是test_chromosome
chromStart(177): The starting position of the feature in the
chromosome or scaffold. The first base in a chromosome is numbered
0.
chromosome上feature(在这里就是junction)的起始位置
chromEnd(400): The ending position of the feature in the chromosome
or scaffold. The chromEnd base is not included in the display of
the feature. For example, the first 100 bases of a chromosome are
defined as chromStart=0, chromEnd=100, and span the bases numbered
0-99.
对应的终止位置
name(JUNC00000001): Defines the name of the BED line.
score(61): A score between 0 and 1000.
strand(+): Defines the strand: either '+' or '-'.
thickStart(177): The starting position at which the feature is
drawn thickly (for example, the start codon in gene
displays).
thickEnd(400): The ending position at which the feature is drawn
thickly (for example, the stop codon in gene displays).
itemRgb(255,0,0): An RGB value of the form R,G,B (e.g. 255,0,0). If
the track line itemRgb attribute is set to "On", this RBG value
will determine the display color of the data contained in this BED
line. NOTE: It is recommended that a simple color scheme (eight
colors or less) be used with this attribute to avoid overwhelming
the color resources of the Genome Browser and your Internet
browser.
blockCount(2): The number of blocks (exons) in the BED line.
外显子个数,2个
blockSizes(73,50): A comma-separated list of the block sizes. The
number of items in this list should correspond to
blockCount.外显子大小,两个数字用逗号隔开
blockStarts(0,173): A comma-separated list of block starts. All of
the blockStart positions should be calculated relative to
chromStart. The number of items in this list should correspond to
blockCount.
外显子起始位置,不从chromosome的起始开始,而是从chromStart开始数,也就是junction的起始位置开始计算,第一个碱基是0
用bed_to_junc转换后得到.juncs其中只保留了junction中intron的部分
cat new_list.juncs
test_chromosome 249 350 + test_chromosome 399 500 +
<chrom> <left> <right> <+/->
http://genome.ucsc.edu/FAQ/FAQformat.html#format1
http://www.ensembl.org/info/website/upload/bed.html
http://zhongguozhanying.blog.163.com/blog/static/21618709520135484450746/
http://blog.sina.com.cn/s/blog_7cffd14001012rtd.html
https://github.com/WormBase/wormbase-pipeline/blob/master/scripts/align_RNASeq.pl