正文
Query ERROR: caught BioMart::Exception::Usage: Attributes from multiple attribute pages are not allowed
就如报错所说, 来源于多个attribute pages 的attributes 被设置.
举个例子:
我有一个 exon ,其id为 ENSE00001706048, 查询其对应的基因id:
## 设置数据库和数据集
human <- useEnsembl(biomart = "genes", dataset = "hsapiens_gene_ensembl", mirror = "asia")
results <- getBM(
attributes= c("ensembl_gene_id", "external_gene_name", "ensembl_exon_id"),
filters=c("ensembl_exon_id"),
values="ENSE00001706048", mart=human)
> results
ensembl_gene_id external_gene_name ensembl_exon_id
1 ENSG00000188554 NBR1 ENSE00001706048
当我们还想,知道exon 的起始,和终止位置时, 加上两个attributes:
results <- getBM(
attributes= c("ensembl_gene_id", "external_gene_name","ensembl_exon_id",
"exon_chrom_start", "exon_chrom_end"),
filters=c("ensembl_exon_id"),
values="ENSE00001706048", mart=human)
也能正常得出我们想要的结果:
> results
ensembl_gene_id external_gene_name ensembl_exon_id exon_chrom_start exon_chrom_end
1 ENSG00000188554 NBR1 ENSE00001706048 43200167 43200608
进一步,若还想知道gene 对应的GO term有哪些, 尝试添加go_id, 这个attribute。
results <- getBM(
attributes= c("ensembl_gene_id", "external_gene_name", "ensembl_exon_id",
"exon_chrom_start", "exon_chrom_end", "go_id"),
filters=c("ensembl_exon_id"),
values="ENSE00001706048", mart=human)
很遗憾,它报错了
Error in .processResults(postRes, mart = mart, hostURLsep = sep, fullXmlQuery = fullXmlQuery, :
Query ERROR: caught BioMart::Exception::Usage: Attributes from multiple attribute pages are not allowed
我们查看下我们设置的attributes,
e_attrs <- c("ensembl_gene_id", "external_gene_name", "ensembl_exon_id", "exon_chrom_start", "exon_chrom_end", "go_id")
listAttributes(human)[listAttributes(human)$name %in% e_attrs, ]
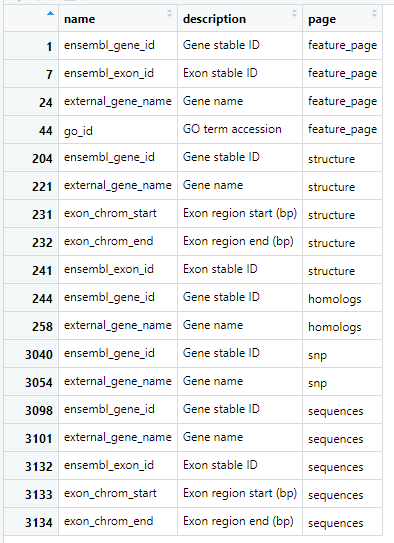
"ensembl_gene_id", "external_gene_name","ensembl_exon_id", "exon_chrom_start", "exon_chrom_end" 都属于structure 这个page, 而feature_page 这个page下,有"go_id", 但没有"exon_chrom_start", "exon_chrom_end"。
所以就如报错所说, 来源于多个attribute pages 的attributes 被设置. "exon_chrom_start", "exon_chrom_end" 和"go_id" 混在一起报错了。
解决方法
分开查询,然后合并了。
results1 <- getBM(
attributes= c("ensembl_gene_id", "external_gene_name", "ensembl_exon_id",
"exon_chrom_start", "exon_chrom_end"),
filters=c("ensembl_exon_id"),
values="ENSE00001706048", mart=human)
results2 <- getBM(
attributes= c("ensembl_gene_id", "external_gene_name", "ensembl_exon_id", "go_id"),
filters=c("ensembl_exon_id"),
values="ENSE00001706048", mart=human)
merge(results1, results2)
> merge(results1, results2)
ensembl_gene_id external_gene_name ensembl_exon_id exon_chrom_start exon_chrom_end go_id
1 ENSG00000188554 NBR1 ENSE00001706048 43200167 43200608 GO:0008270
2 ENSG00000188554 NBR1 ENSE00001706048 43200167 43200608 GO:0005515
3 ENSG00000188554 NBR1 ENSE00001706048 43200167 43200608 GO:0043130
4 ENSG00000188554 NBR1 ENSE00001706048 43200167 43200608 GO:0000407
5 ENSG00000188554 NBR1 ENSE00001706048 43200167 43200608 GO:0016236
...........
23 ENSG00000188554 NBR1 ENSE00001706048 43200167 43200608 GO:0051019
24 ENSG00000188554 NBR1 ENSE00001706048 43200167 43200608 GO:0032872
25 ENSG00000188554 NBR1 ENSE00001706048 43200167 43200608 GO:0005758
其他
listAttributes 函数可以列出,可查询返回的attributes ,listFilters可以列出可以用于筛选的attributes
> ensembl <- useEnsembl(biomart = "genes", dataset = "hsapiens_gene_ensembl", mirror = "asia")
> listAttributes(ensembl)
name description page
1 ensembl_gene_id Gene stable ID feature_page
2 ensembl_gene_id_version Gene stable ID version feature_page
3 ensembl_transcript_id Transcript stable ID feature_page
4 ensembl_transcript_id_version Transcript stable ID version feature_page
5 ensembl_peptide_id Protein stable ID feature_page
6 ensembl_peptide_id_version Protein stable ID version feature_page
..........
..........
> listFilters(ensembl)
name description
1 chromosome_name Chromosome/scaffold name
2 start Start
3 end End
4 strand Strand
5 chromosomal_region e.g. 1:100:10000:-1, 1:100000:200000:1
......
.....
以及biomaRt, 是个好东西,就是经常提醒我请求尝试超时...
Error in curl::curl_fetch_memory(url, handle = handle) :
Timeout was reached: [asia.ensembl.org:443] Connection timed out after 10001 milliseconds
参考
https://bioconductor.org/packages/release/bioc/vignettes/biomaRt/inst/doc/accessing_ensembl.html
https://support.bioconductor.org/p/33414/