Chapter 3 - Regression Models
Segment 3 - Logistic regression
Logistic Regression
Logistic regression is a simple machine learning method you can use to predict the value of a numeric categorical variable based on its relationship with predictor variables.
Logistic Regression Use Cases
- Customer Churn Prediction
- Employee Attrition Modeling
- Hazardous Event Prediction
- Purchase Propensity vs. Ad Spend Analysis
Logistic Regression Assumptions
- Data is free of missing values
- The predictant variable is binary(that is, it only accepts two values) or ordinal (that is, a categorical variable with ordered values)
- All predictors are independent of each other
- There are at least 50 observations per predictor variable (to ensure reliable results)
import numpy as np
import pandas as pd
import seaborn as sb
import matplotlib.pyplot as plt
import sklearn
from pandas import Series, DataFrame
from pylab import rcParams
from sklearn import preprocessing
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_val_predict
from sklearn import metrics
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
from sklearn.metrics import precision_score, recall_score
%matplotlib inline
rcParams['figure.figsize'] = 5, 4
sb.set_style('whitegrid')
Logistic regression on the titanic dataset
address = '~/Data/titanic-training-data.csv'
titanic_training = pd.read_csv(address)
titanic_training.columns = ['PassengerId','Survived','Pclass','Name','Sex','Age','SibSp','Parch','Ticket','Fare','Cabin','Embarked']
print(titanic_training.head())
PassengerId Survived Pclass
0 1 0 3
1 2 1 1
2 3 1 3
3 4 1 1
4 5 0 3
Name Sex Age SibSp
0 Braund, Mr. Owen Harris male 22.0 1
1 Cumings, Mrs. John Bradley (Florence Briggs Th... female 38.0 1
2 Heikkinen, Miss. Laina female 26.0 0
3 Futrelle, Mrs. Jacques Heath (Lily May Peel) female 35.0 1
4 Allen, Mr. William Henry male 35.0 0
Parch Ticket Fare Cabin Embarked
0 0 A/5 21171 7.2500 NaN S
1 0 PC 17599 71.2833 C85 C
2 0 STON/O2. 3101282 7.9250 NaN S
3 0 113803 53.1000 C123 S
4 0 373450 8.0500 NaN S
print(titanic_training.info())
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 891 non-null int64
1 Survived 891 non-null int64
2 Pclass 891 non-null int64
3 Name 891 non-null object
4 Sex 891 non-null object
5 Age 714 non-null float64
6 SibSp 891 non-null int64
7 Parch 891 non-null int64
8 Ticket 891 non-null object
9 Fare 891 non-null float64
10 Cabin 204 non-null object
11 Embarked 889 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 83.7+ KB
None
VARIABLE DESCRIPTIONS
Survived - Survival (0 = No; 1 = Yes)
Pclass - Passenger Class (1 = 1st; 2 = 2nd; 3 = 3rd)
Name - Name
Sex - Sex
Age - Age
SibSp - Number of Siblings/Spouses Aboard
Parch - Number of Parents/Children Aboard
Ticket - Ticket Number
Fare - Passenger Fare (British pound)
Cabin - Cabin
Embarked - Port of Embarkation (C = Cherbourg, France; Q = Queenstown, UK; S = Southampton - Cobh, Ireland)
Checking that your target variable is binary
sb.countplot(x='Survived', data=titanic_training, palette='hls')
<matplotlib.axes._subplots.AxesSubplot at 0x7ff6fe9b95c0>
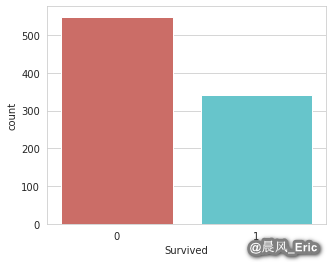
Checking for missing values
titanic_training.isnull().sum()
PassengerId 0
Survived 0
Pclass 0
Name 0
Sex 0
Age 177
SibSp 0
Parch 0
Ticket 0
Fare 0
Cabin 687
Embarked 2
dtype: int64
titanic_training.describe()
|
PassengerId |
Survived |
Pclass |
Age |
SibSp |
Parch |
Fare |
| count |
891.000000 |
891.000000 |
891.000000 |
714.000000 |
891.000000 |
891.000000 |
891.000000 |
| mean |
446.000000 |
0.383838 |
2.308642 |
29.699118 |
0.523008 |
0.381594 |
32.204208 |
| std |
257.353842 |
0.486592 |
0.836071 |
14.526497 |
1.102743 |
0.806057 |
49.693429 |
| min |
1.000000 |
0.000000 |
1.000000 |
0.420000 |
0.000000 |
0.000000 |
0.000000 |
| 25% |
223.500000 |
0.000000 |
2.000000 |
20.125000 |
0.000000 |
0.000000 |
7.910400 |
| 50% |
446.000000 |
0.000000 |
3.000000 |
28.000000 |
0.000000 |
0.000000 |
14.454200 |
| 75% |
668.500000 |
1.000000 |
3.000000 |
38.000000 |
1.000000 |
0.000000 |
31.000000 |
| max |
891.000000 |
1.000000 |
3.000000 |
80.000000 |
8.000000 |
6.000000 |
512.329200 |
Taking care of missing values
Dropping missing values
So let's just go ahead and drop all the variables that aren't relevant for predicting survival. We should at least keep the following:
- Survived - This variable is obviously relevant.
- Pclass - Does a passenger's class on the boat affect their survivability?
- Sex - Could a passenger's gender impact their survival rate?
- Age - Does a person's age impact their survival rate?
- SibSp - Does the number of relatives on the boat (that are siblings or a spouse) affect a person survivability? Probability
- Parch - Does the number of relatives on the boat (that are children or parents) affect a person survivability? Probability
- Fare - Does the fare a person paid effect his survivability? Maybe - let's keep it.
- Embarked - Does a person's point of embarkation matter? It depends on how the boat was filled... Let's keep it.
What about a person's name, ticket number, and passenger ID number? They're irrelavant for predicting survivability. And as you recall, the cabin variable is almost all missing values, so we can just drop all of these.
titanic_data = titanic_training.drop(['Name','Ticket','Cabin'], axis=1)
titanic_data.head()
|
PassengerId |
Survived |
Pclass |
Sex |
Age |
SibSp |
Parch |
Fare |
Embarked |
| 0 |
1 |
0 |
3 |
male |
22.0 |
1 |
0 |
7.2500 |
S |
| 1 |
2 |
1 |
1 |
female |
38.0 |
1 |
0 |
71.2833 |
C |
| 2 |
3 |
1 |
3 |
female |
26.0 |
0 |
0 |
7.9250 |
S |
| 3 |
4 |
1 |
1 |
female |
35.0 |
1 |
0 |
53.1000 |
S |
| 4 |
5 |
0 |
3 |
male |
35.0 |
0 |
0 |
8.0500 |
S |
Imputing missing values
sb.boxplot(x='Parch', y='Age', data=titanic_data, palette='hls')
<matplotlib.axes._subplots.AxesSubplot at 0x7ff6fc140828>
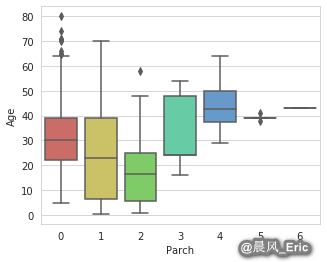
Parch_groups = titanic_data.groupby(titanic_data['Parch'])
Parch_groups.mean()
|
PassengerId |
Survived |
Pclass |
Age |
SibSp |
Fare |
| Parch |
|
|
|
|
|
|
| 0 |
445.255162 |
0.343658 |
2.321534 |
32.178503 |
0.237463 |
25.586774 |
| 1 |
465.110169 |
0.550847 |
2.203390 |
24.422000 |
1.084746 |
46.778180 |
| 2 |
416.662500 |
0.500000 |
2.275000 |
17.216912 |
2.062500 |
64.337604 |
| 3 |
579.200000 |
0.600000 |
2.600000 |
33.200000 |
1.000000 |
25.951660 |
| 4 |
384.000000 |
0.000000 |
2.500000 |
44.500000 |
0.750000 |
84.968750 |
| 5 |
435.200000 |
0.200000 |
3.000000 |
39.200000 |
0.600000 |
32.550000 |
| 6 |
679.000000 |
0.000000 |
3.000000 |
43.000000 |
1.000000 |
46.900000 |
def age_approx(cols):
Age = cols[0]
Parch = cols[1]
if pd.isnull(Age):
if Parch == 0:
return 32
elif Parch == 1:
return 24
elif Parch == 2:
return 17
elif Parch == 3:
return 33
elif Parch == 4:
return 45
else:
return 30
else:
return Age
titanic_data['Age'] = titanic_data[['Age','Parch']].apply(age_approx, axis=1)
titanic_data.isnull().sum()
PassengerId 0
Survived 0
Pclass 0
Sex 0
Age 0
SibSp 0
Parch 0
Fare 0
Embarked 2
dtype: int64
titanic_data.dropna(inplace=True)
titanic_data.reset_index(inplace=True, drop=True)
print(titanic_data.info())
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 889 entries, 0 to 888
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 889 non-null int64
1 Survived 889 non-null int64
2 Pclass 889 non-null int64
3 Sex 889 non-null object
4 Age 889 non-null float64
5 SibSp 889 non-null int64
6 Parch 889 non-null int64
7 Fare 889 non-null float64
8 Embarked 889 non-null object
dtypes: float64(2), int64(5), object(2)
memory usage: 62.6+ KB
None
Converting categorical variables to a dummy indicators
from sklearn.preprocessing import LabelEncoder
label_encoder = LabelEncoder()
gender_cat = titanic_data['Sex']
gender_encoded = label_encoder.fit_transform(gender_cat)
gender_encoded[0:5]
array([1, 0, 0, 0, 1])
titanic_data.head()
|
PassengerId |
Survived |
Pclass |
Sex |
Age |
SibSp |
Parch |
Fare |
Embarked |
| 0 |
1 |
0 |
3 |
male |
22.0 |
1 |
0 |
7.2500 |
S |
| 1 |
2 |
1 |
1 |
female |
38.0 |
1 |
0 |
71.2833 |
C |
| 2 |
3 |
1 |
3 |
female |
26.0 |
0 |
0 |
7.9250 |
S |
| 3 |
4 |
1 |
1 |
female |
35.0 |
1 |
0 |
53.1000 |
S |
| 4 |
5 |
0 |
3 |
male |
35.0 |
0 |
0 |
8.0500 |
S |
# 1 = male / 0 = female
gender_DF = pd.DataFrame(gender_encoded, columns=['male_gender'])
gender_DF.head()
|
male_gender |
| 0 |
1 |
| 1 |
0 |
| 2 |
0 |
| 3 |
0 |
| 4 |
1 |
embarked_cat = titanic_data['Embarked']
embarked_encoded = label_encoder.fit_transform(embarked_cat)
embarked_encoded[0:100]
array([2, 0, 2, 2, 2, 1, 2, 2, 2, 0, 2, 2, 2, 2, 2, 2, 1, 2, 2, 0, 2, 2,
1, 2, 2, 2, 0, 2, 1, 2, 0, 0, 1, 2, 0, 2, 0, 2, 2, 0, 2, 2, 0, 0,
1, 2, 1, 1, 0, 2, 2, 2, 0, 2, 0, 2, 2, 0, 2, 2, 0, 2, 2, 0, 0, 2,
2, 2, 2, 2, 2, 2, 0, 2, 2, 2, 2, 2, 2, 2, 2, 1, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 0, 0, 2, 2, 2])
from sklearn.preprocessing import OneHotEncoder
binary_encoder = OneHotEncoder(categories='auto')
embarked_1hot = binary_encoder.fit_transform(embarked_encoded.reshape(-1,1))
embarked_1hot_mat = embarked_1hot.toarray()
embarked_DF = pd.DataFrame(embarked_1hot_mat, columns = ['C','Q','S'])
embarked_DF.head()
|
C |
Q |
S |
| 0 |
0.0 |
0.0 |
1.0 |
| 1 |
1.0 |
0.0 |
0.0 |
| 2 |
0.0 |
0.0 |
1.0 |
| 3 |
0.0 |
0.0 |
1.0 |
| 4 |
0.0 |
0.0 |
1.0 |
titanic_data.drop(['Sex','Embarked'], axis=1, inplace=True)
titanic_data.head()
|
PassengerId |
Survived |
Pclass |
Age |
SibSp |
Parch |
Fare |
| 0 |
1 |
0 |
3 |
22.0 |
1 |
0 |
7.2500 |
| 1 |
2 |
1 |
1 |
38.0 |
1 |
0 |
71.2833 |
| 2 |
3 |
1 |
3 |
26.0 |
0 |
0 |
7.9250 |
| 3 |
4 |
1 |
1 |
35.0 |
1 |
0 |
53.1000 |
| 4 |
5 |
0 |
3 |
35.0 |
0 |
0 |
8.0500 |
titanic_dmy = pd.concat([titanic_data, gender_DF, embarked_DF], axis=1, verify_integrity=True).astype(float)
titanic_dmy[0:5]
|
PassengerId |
Survived |
Pclass |
Age |
SibSp |
Parch |
Fare |
male_gender |
C |
Q |
S |
| 0 |
1.0 |
0.0 |
3.0 |
22.0 |
1.0 |
0.0 |
7.2500 |
1.0 |
0.0 |
0.0 |
1.0 |
| 1 |
2.0 |
1.0 |
1.0 |
38.0 |
1.0 |
0.0 |
71.2833 |
0.0 |
1.0 |
0.0 |
0.0 |
| 2 |
3.0 |
1.0 |
3.0 |
26.0 |
0.0 |
0.0 |
7.9250 |
0.0 |
0.0 |
0.0 |
1.0 |
| 3 |
4.0 |
1.0 |
1.0 |
35.0 |
1.0 |
0.0 |
53.1000 |
0.0 |
0.0 |
0.0 |
1.0 |
| 4 |
5.0 |
0.0 |
3.0 |
35.0 |
0.0 |
0.0 |
8.0500 |
1.0 |
0.0 |
0.0 |
1.0 |
Checking for independence between features
sb.heatmap(titanic_dmy.corr())
<matplotlib.axes._subplots.AxesSubplot at 0x7ff6fc071dd8>
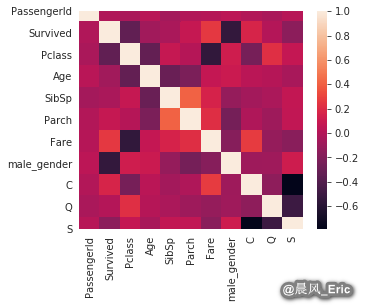
titanic_dmy.drop(['Fare','Pclass'], axis=1, inplace=True)
titanic_dmy.head()
|
PassengerId |
Survived |
Age |
SibSp |
Parch |
male_gender |
C |
Q |
S |
| 0 |
1.0 |
0.0 |
22.0 |
1.0 |
0.0 |
1.0 |
0.0 |
0.0 |
1.0 |
| 1 |
2.0 |
1.0 |
38.0 |
1.0 |
0.0 |
0.0 |
1.0 |
0.0 |
0.0 |
| 2 |
3.0 |
1.0 |
26.0 |
0.0 |
0.0 |
0.0 |
0.0 |
0.0 |
1.0 |
| 3 |
4.0 |
1.0 |
35.0 |
1.0 |
0.0 |
0.0 |
0.0 |
0.0 |
1.0 |
| 4 |
5.0 |
0.0 |
35.0 |
0.0 |
0.0 |
1.0 |
0.0 |
0.0 |
1.0 |
Checking that your dataset size is sufficient
titanic_dmy.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 889 entries, 0 to 888
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 889 non-null float64
1 Survived 889 non-null float64
2 Age 889 non-null float64
3 SibSp 889 non-null float64
4 Parch 889 non-null float64
5 male_gender 889 non-null float64
6 C 889 non-null float64
7 Q 889 non-null float64
8 S 889 non-null float64
dtypes: float64(9)
memory usage: 62.6 KB
X_train, X_test, y_train, y_test = train_test_split(titanic_dmy.drop('Survived',axis=1),
titanic_dmy['Survived'],test_size=0.2,
random_state=200)
print(X_train.shape)
print(y_train.shape)
(711, 8)
(711,)
X_train[0:5]
|
PassengerId |
Age |
SibSp |
Parch |
male_gender |
C |
Q |
S |
| 719 |
721.0 |
6.0 |
0.0 |
1.0 |
0.0 |
0.0 |
0.0 |
1.0 |
| 165 |
167.0 |
24.0 |
0.0 |
1.0 |
0.0 |
0.0 |
0.0 |
1.0 |
| 879 |
882.0 |
33.0 |
0.0 |
0.0 |
1.0 |
0.0 |
0.0 |
1.0 |
| 451 |
453.0 |
30.0 |
0.0 |
0.0 |
1.0 |
1.0 |
0.0 |
0.0 |
| 181 |
183.0 |
9.0 |
4.0 |
2.0 |
1.0 |
0.0 |
0.0 |
1.0 |
Deploying and evaluating the model
LogReg = LogisticRegression(solver='liblinear')
LogReg.fit(X_train, y_train)
LogisticRegression(solver='liblinear')
y_pred = LogReg.predict(X_test)
Model Evaluation
Classification report without cross-validation
print(classification_report(y_test, y_pred))
precision recall f1-score support
0.0 0.83 0.88 0.85 109
1.0 0.79 0.71 0.75 69
accuracy 0.81 178
macro avg 0.81 0.80 0.80 178
weighted avg 0.81 0.81 0.81 178
K-fold cross-validation & confusion matrices
y_train_pred = cross_val_predict(LogReg, X_train, y_train, cv=5)
confusion_matrix(y_train, y_train_pred)
array([[377, 63],
[ 91, 180]])
precision_score(y_train, y_train_pred)
0.7407407407407407
Make a test prediction
titanic_dmy[863:864]
|
PassengerId |
Survived |
Age |
SibSp |
Parch |
male_gender |
C |
Q |
S |
| 863 |
866.0 |
1.0 |
42.0 |
0.0 |
0.0 |
0.0 |
0.0 |
0.0 |
1.0 |
test_passenger = np.array([866,40,0,0,0,0,0,1]).reshape(1,-1)
print(LogReg.predict(test_passenger))
print(LogReg.predict_proba(test_passenger))
[1.]
[[0.26351831 0.73648169]]


