问题:
- 为什么组间方差加组内方差一定等于总方差?如何从数学上理解。PPT里有证明,引入一个中间项就行。
- 方差分析、协方差分析和回归分析有什么联系?
- 什么是F分布?Fisher的独创,理解不了F分布就不可能真正理解方差分析。
- 方差分析,就是要分析方差的来源!
我们把组内方差看做是随机误差;组间差组成比较复杂:包含了随机误差、系统误差。
回忆一下:
卡方分布就是多个标准正态分布变量平方的和,自由度是其唯一的参数。(为什么当自由度为3时,卡方分布的形状就变了,和三体问题有关吗?)
F分布就是两个不同卡方分布的比的分布,自由度是其唯一的参数(两个自由度而已)。
方差分析假设随机误差是服从正态分布的,那么我们假设组内和组间无差异,很自然就转换到了F分布。
那就连t分布一起回顾吧!t就是学生的意思,著名的t-SNE也是基于t分布的,t分布和正态分布形状基本是一样的,当t分布唯一的参数自由度大于30时,t分布就趋近于正态分布了。普通的z分布底下除的是总体标准差,t分布底下除的是样本标准差。t分布的自由度就是抽样分布中的sample size,根据中心极限定理,sample size越大,抽样分布的均值就越趋近于正态分布。【YouTube上有个视频讲得非常清楚】
原理
比较两组(小样本)就用t-test,比较三组及以上就用ANOVA。注意:我们默认说的都是one way ANOVA,也就是对group的分类标准只有一个,比如case和control(ABCD多组),two way就是分类标准有多个,比如case or control,male or femal。
方差分析的核心原理:组内方差和组间方差是否有明显的差异,用的F统计量,F分布有两个参数,也就是两个自由度参数。
方差分析会给一个总的显著性结果,及组内和组间是否有显著差异。显著了需要再做两两比较。

R实例
my_data <- PlantGrowth
# Show a random sample
set.seed(1234)
dplyr::sample_n(my_data, 10)
# Show the levels
levels(my_data$group)
my_data$group <- ordered(my_data$group,
levels = c("ctrl", "trt1", "trt2"))
library(dplyr)
group_by(my_data, group) %>%
summarise(
count = n(),
mean = mean(weight, na.rm = TRUE),
sd = sd(weight, na.rm = TRUE)
)
# Box plots
# ++++++++++++++++++++
# Plot weight by group and color by group
library("ggpubr")
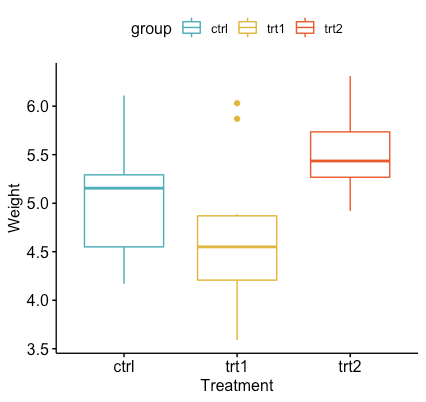
ggboxplot(my_data, x = "group", y = "weight",
color = "group", palette = c("#00AFBB", "#E7B800", "#FC4E07"),
order = c("ctrl", "trt1", "trt2"),
ylab = "Weight", xlab = "Treatment")
# Mean plots
# ++++++++++++++++++++
# Plot weight by group
# Add error bars: mean_se
# (other values include: mean_sd, mean_ci, median_iqr, ....)
library("ggpubr")
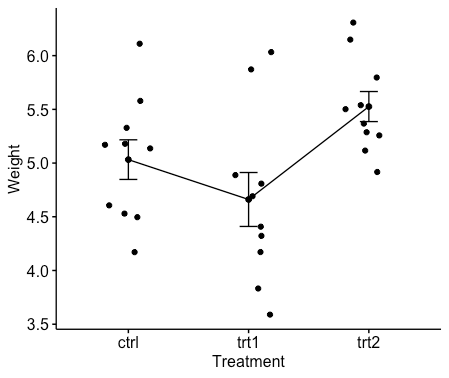
ggline(my_data, x = "group", y = "weight",
add = c("mean_se", "jitter"),
order = c("ctrl", "trt1", "trt2"),
ylab = "Weight", xlab = "Treatment")
# Box plot
boxplot(weight ~ group, data = my_data,
xlab = "Treatment", ylab = "Weight",
frame = FALSE, col = c("#00AFBB", "#E7B800", "#FC4E07"))
# plotmeans
library("gplots")
plotmeans(weight ~ group, data = my_data, frame = FALSE,
xlab = "Treatment", ylab = "Weight",
main="Mean Plot with 95% CI")
# Compute the analysis of variance
res.aov <- aov(weight ~ group, data = my_data)
# Summary of the analysis
summary(res.aov)
# In one-way ANOVA test, a significant p-value indicates that some of the group means are different,
# but we don’t know which pairs of groups are different.
TukeyHSD(res.aov)
进阶
HSD
general linear hypothesis tests
repalce by Pairewise t-test under BH adjust
test validity
One-Way vs Two-Way ANOVA: Differences, Assumptions and Hypotheses