1、测试数据下载
链接:https://pan.baidu.com/s/1EfffExvtxZYI1QLuxUZQ_g
提取码:5wfe
数据为plink 格式数据test.map、test.ped ;
一共包含三个品种,DOR、GMM、SUN各20个样本。
2、使用plink将测试数据转化为二进制:
plink --file test --make-bed --out test
生成文件结果如下:
3、运行admixture软件
for i in $(seq 5);do admixture --cv test.bed $i|tee $i.log;done
命令解释:
seq 5 : 生成1 2 3 4 5自然数,为假设的祖先数目
--cv:进行交叉错误率验证
tee $i.log: 记录命令运行过程
运行结果如下:
4、查看交叉错误率验证结果
命令如下:
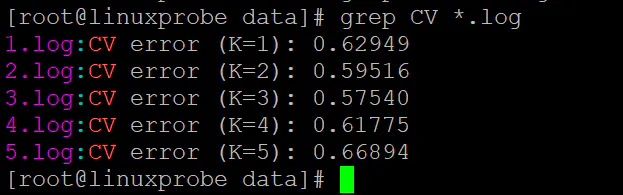
grep CV *.log
结果如下,可见k等于3时交叉错误率最小:
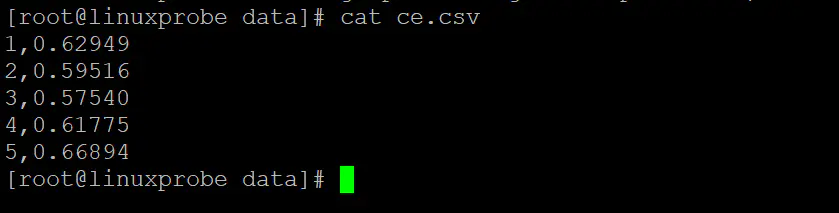
对交叉验证错误进行可视化,首先生成可读文件:
grep CV *.log|awk '{print NR","$NF}' >ce.csv
利用R进行绘图,命令如下:
mydat <- read.csv("ce.csv",header = F,as.is = T,stringsAsFactors = F)
png("ce.png",width = 7000,height = 7000,pointsize = 160)
plot(mydat$V1,mydat$V2,type = "o",col = "red",pch = 19,xlab = "K",ylab = "Cross-validation error", lwd.ticks=10,font.axis=2,cex.lab=1.2,lwd = 10,cex.axis=1.2,font.lab=2)
box(which="plot",col="black",lwd=18)
dev.off()
绘图结果:
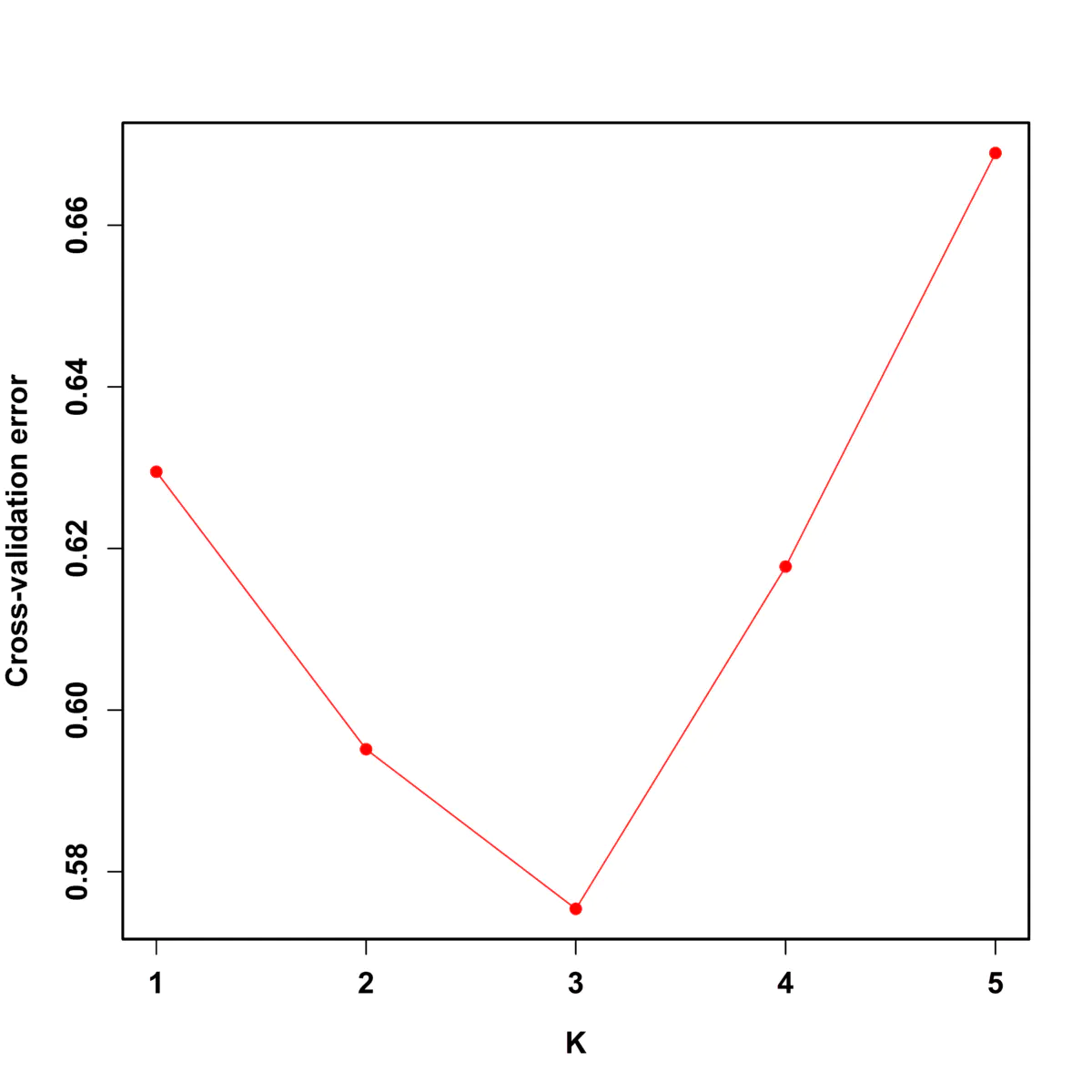
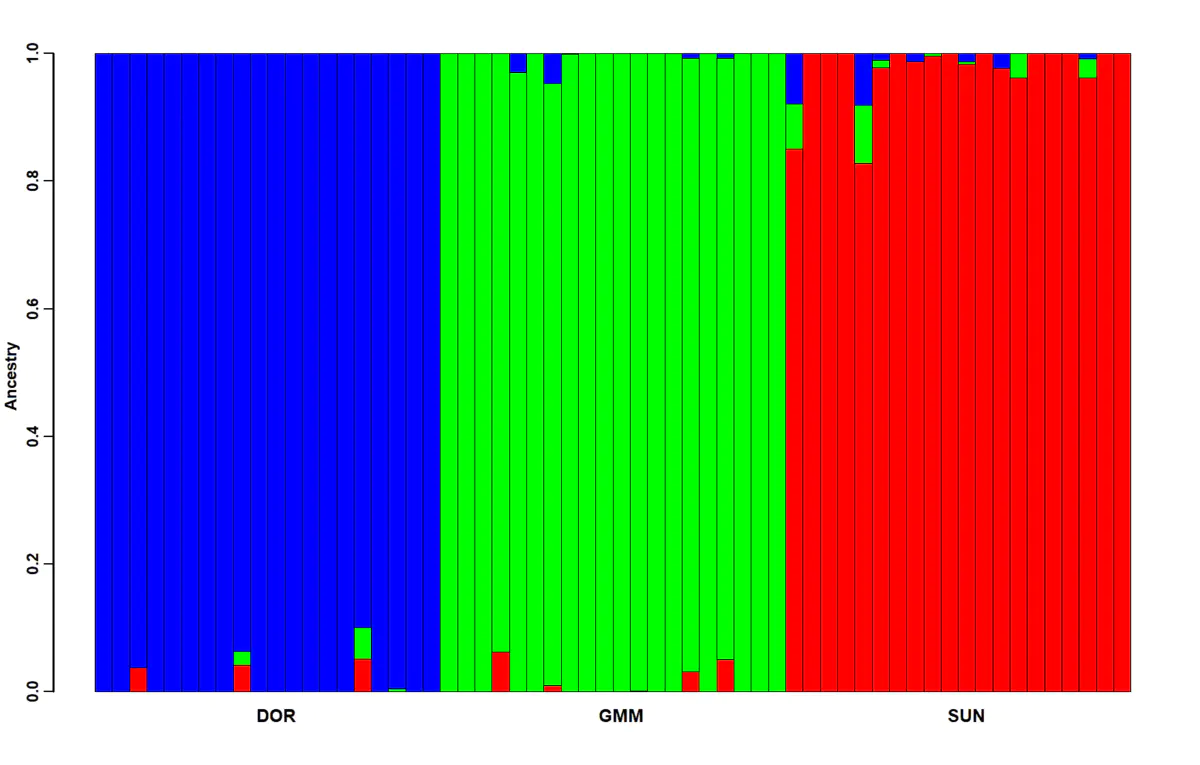
5、利用k等于3的结果进行绘图,命令如下:
mydat=read.table("test.3.Q")
barplot(t(as.matrix(mydat)), col=rainbow(3),cex.axis=1.5,font.axis = 2, lwd.ticks=3,yaxt="n",lwd.axis = 5,space = 0,xaxt = "n")
axis(1, at=c(10.5,30.5,50.5),labels = c("DOR","GMM","SUN"), tck = -0.015,tick = F, cex.axis=1.6,font = 2, lwd.ticks = 2.5,lwd = 2.7)
axis(2, at=seq(0,1.0,0.2), tck = -0.015, cex.axis=1.45,font = 2, lwd.ticks = 2.5,lwd = ) mtext(side = 2,"Ancestry",cex = 1.5,font = 2,line = 2.7)
绘图结果: