作者|THILAKADIBOINA
编译|Flin
来源|analyticsvidhya
介绍
本文介绍了生成性对抗网络(Generative attersarial Networks,GAN)的使用,这是一种对真实的Covid-19数据进行过采样的技术,用于预测死亡率。这个故事让我们更好地理解数据准备步骤(如处理不平衡的数据)如何提高模型性能。
本文的数据和核心模型来自Celestine Iwendi、Ali Kashif Bashir、Atharva Peshkar最近的一项研究(2020年7月)“使用增强随机森林算法预测COVID-19患者健康”。本研究使用ADABOST模型增强的随机森林算法预测个体患者的死亡率,准确率为94%。本文考虑相同的模型和模型参数,明确分析了采用基于GAN的过采样技术对现有模型的改进。
对于有抱负的数据科学家来说,学习良好实践的最好方法之一就是参加不同论坛上的黑客竞赛,比如Vidhya、Kaggle或其他论坛。
此外,从这些论坛或出版的研究出版物中获取已解决的案例和数据;了解他们的方法,并尝试通过额外的步骤来提高准确性或减少误差。这将形成一个坚实的基础,使我们能够深入思考我们在数据科学价值链中所学的其他技术的应用。
研究中使用的数据是用222个病人的13个特征来训练的。数据有偏差,159例(72%)属于“0”类或“已恢复”类。由于其偏差性质,各种欠采样/过采样技术可应用于数据。偏态数据的问题会导致预测模型的过度拟合。
为了克服这一局限性,许多研究采用过采样方法来平衡数据集,从而得到更精确的模型训练。过采样是一种通过增加少数数据中的样本数量来补偿数据集不平衡的技术。
常规方法包括随机过采样(ROS)、合成少数过采样技术(SMOTE)等。有关使用常规方法处理不平衡类的更多信息,请参阅:
最近,一种基于对抗性学习概念的生成性网络的机器学习模型被提出,即生成性对抗性网络。生成性对抗网络(Generative atterial Networks,GAN)的特点使其较易应用于过采样研究,因为基于对抗训练的神经网络的性质允许生成与原始数据相似的人工数据。基于生成性对抗网络的过采样克服了传统方法(如过拟合)的局限性,允许建立一个高精度的不平衡数据预测模型。
如何生成合成数据?
两个神经网络相互竞争,学习目标分布并生成人工数据
发生器网络G:模拟训练样本欺骗鉴别器
判别网络D:判别训练样本和生成样本
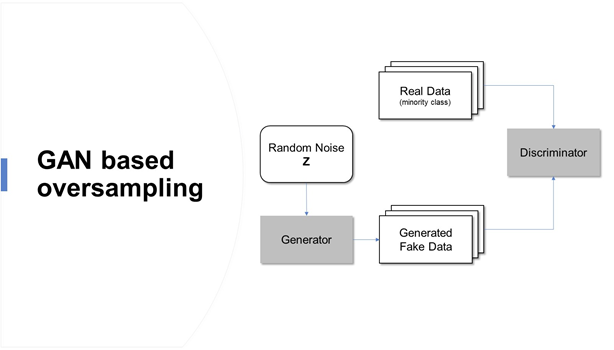
生成性对抗性网络是基于博弈论的场景,其中生成网络必须与对手竞争。随着GAN学会模拟数据的分布,它被应用于各个领域,如音乐、视频和自然语言,最近还用于处理不平衡的数据问题。
研究中使用的数据和基本模型可以在这里找到
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import tensorflow as tf
from keras.layers import Input, Dense, Reshape, Flatten, Dropout, BatchNormalization, Embedding
from keras.layers.advanced_activations import LeakyReLU
from keras.layers.merge import concatenate
from keras.models import Sequential, Model
from keras.optimizers import Adam
from keras.utils import to_categorical
from keras.layers.advanced_activations import LeakyReLU
from keras.utils.vis_utils import plot_model
from sklearn.preprocessing import MinMaxScaler, OneHotEncoder, LabelEncoder
import scipy.stats
import datetime as dt
import pydot
import warnings
warnings.filterwarnings("ignore")
%matplotlib inline
df = pd.read_csv('Covid_Train_Oct32020.csv')
df = df.drop('id',axis=1)
df = df.fillna(np.nan,axis=0)
df['age'] = df['age'].fillna(value=df['age'].mean())
df['sym_on'] = pd.to_datetime(df['sym_on'])
df['hosp_vis'] = pd.to_datetime(df['hosp_vis'])
df['sym_on']= df['sym_on'].map(dt.datetime.toordinal)
df['hosp_vis']= df['hosp_vis'].map(dt.datetime.toordinal)
df['diff_sym_hos']= df['hosp_vis'] - df['sym_on']
df=df.drop(['sym_on', 'hosp_vis'], axis=1)
df['location'] = df['location'].astype(str)
df['country'] = df['country'].astype(str)
df['gender'] = df['gender'].astype(str)
df['vis_wuhan'] = df['vis_wuhan'].astype(str)
df['from_wuhan'] = df['from_wuhan'].astype(str)
df['symptom1'] = df['symptom1'].astype(str)
df['symptom2'] = df['symptom2'].astype(str)
df['symptom3'] = df['symptom3'].astype(str)
df['symptom4'] = df['symptom4'].astype(str)
df['symptom5'] = df['symptom5'].astype(str)
df['symptom6'] = df['symptom6'].astype(str)
df.dtypes
数据说明
| 列 | 描述 | 值(用于分类变量) | 类型 |
| id | 患者编号 | 不适用 | 数字 |
| location | 患者所属的位置 | 遍布全球的多个城市 | 字符串,分类 |
| country | 患者的国家 | 多个国家 | 字符串,分类 |
| gender | 患者性别 | 男,女 | 字符串,分类 |
| age | 患者年龄 | 不适用 | 数字 |
| sym_on | 患者开始注意到症状的日期 | 不适用 | 日期 |
| hosp_vis | 病人去医院的日期 | 不适用 | 日期 |
| vis_wuhan | 患者是否去过中国武汉 | 是(1),否(0) | 数值,分类 |
| from_wuhan | 患者是否属于中国武汉 | 是(1),否(0) | 数值,分类 |
| death | 患者是否因COVID-19而去世 | 是(1),否(0) | 数值,分类 |
| Recov | 患者是否康复 | 是(1),否(0) | 数值,分类 |
| symptom1. symptom2, symptom3, symptom4, symptom5, symptom6 | 患者注意到的症状 | 患者注意到多种症状 | 字符串,分类 |
该研究考虑了11个分类输入特征和2个数字输入特征。目标变量是死亡/恢复。已填充新列“ diff_sym_hos”,以提供当天在医院发现和接受的症状之间的差异。
研究的重点是改善少数类别数据,即死亡== 1,从训练数据中提取了一个子集。子集按类别和数字分开,并传递给GAN模型。
df_minority_data=df.loc[df['death'] == 1]
#Subsetting input features without target variable
df_minority_data_withouttv=df_minority_data.loc[:, df_minority_data.columns != 'death']
numerical_df = df_minority_data_withouttv.select_dtypes("number")
categorical_df = df_minority_data_withouttv.select_dtypes("object")
scaling = MinMaxScaler()
numerical_df_rescaled = scaling.fit_transform(numerical_df)
get_dummy_df = pd.get_dummies(categorical_df)
#Seperating Each Category
location_dummy_col = [col for col in get_dummy_df.columns if 'location' in col]
location_dummy = get_dummy_df[location_dummy_col]
country_dummy_col = [col for col in get_dummy_df.columns if 'country' in col]
country_dummy = get_dummy_df[country_dummy_col]
gender_dummy_col = [col for col in get_dummy_df.columns if 'gender' in col]
gender_dummy = get_dummy_df[gender_dummy_col]
vis_wuhan_dummy_col = [col for col in get_dummy_df.columns if 'vis_wuhan' in col]
vis_wuhan_dummy = get_dummy_df[vis_wuhan_dummy_col]
from_wuhan_dummy_col = [col for col in get_dummy_df.columns if 'from_wuhan' in col]
from_wuhan_dummy = get_dummy_df[from_wuhan_dummy_col]
symptom1_dummy_col = [col for col in get_dummy_df.columns if 'symptom1' in col]
symptom1_dummy = get_dummy_df[symptom1_dummy_col]
symptom2_dummy_col = [col for col in get_dummy_df.columns if 'symptom2' in col]
symptom2_dummy = get_dummy_df[symptom2_dummy_col]
symptom3_dummy_col = [col for col in get_dummy_df.columns if 'symptom3' in col]
symptom3_dummy = get_dummy_df[symptom3_dummy_col]
symptom4_dummy_col = [col for col in get_dummy_df.columns if 'symptom4' in col]
symptom4_dummy = get_dummy_df[symptom4_dummy_col]
symptom5_dummy_col = [col for col in get_dummy_df.columns if 'symptom5' in col]
symptom5_dummy = get_dummy_df[symptom5_dummy_col]
symptom6_dummy_col = [col for col in get_dummy_df.columns if 'symptom6' in col]
symptom6_dummy = get_dummy_df[symptom6_dummy_col]

定义生成器
生成器从潜在空间获取输入并生成新的合成样本。泄露修正线性单元(LeakyReLU)是在发生器和鉴别器模型中用于处理某些负值的函数。
它使用默认建议值0.2和适当的权重初始化程序“ he_uniform”使用。此外,在不同的层之间使用批处理归一化来标准化来自先前层的激活(零均值和单位方差)并稳定训练过程。
在输出层中,softmax激活函数用于分类变量,而sigmoid 函数用于连续变量。
def define_generator (catsh1,catsh2,catsh3,catsh4,catsh5,catsh6,catsh7,catsh8,catsh9,catsh10,catsh11,numerical):
#Inputting noise from latent space
noise = Input(shape = (70,))
hidden_1 = Dense(8, kernel_initializer = "he_uniform")(noise)
hidden_1 = LeakyReLU(0.2)(hidden_1)
hidden_1 = BatchNormalization(momentum = 0.8)(hidden_1)
hidden_2 = Dense(16, kernel_initializer = "he_uniform")(hidden_1)
hidden_2 = LeakyReLU(0.2)(hidden_2)
hidden_2 = BatchNormalization(momentum = 0.8)(hidden_2)
#Branch 1 for generating location data
branch_1 = Dense(32, kernel_initializer = "he_uniform")(hidden_2)
branch_1 = LeakyReLU(0.2)(branch_1)
branch_1 = BatchNormalization(momentum = 0.8)(branch_1)
branch_1 = Dense(64, kernel_initializer = "he_uniform")(branch_1)
branch_1 = LeakyReLU(0.2)(branch_1)
branch_1 = BatchNormalization(momentum=0.8)(branch_1)
#Output Layer1
branch_1_output = Dense(catsh1, activation = "softmax")(branch_1)
#Likewise, for all remaining 10 categories branches will be defined
#Branch 12 for generating numerical data
branch_12 = Dense(64, kernel_initializer = "he_uniform")(hidden_2)
branch_12 = LeakyReLU(0.2)(branch_3)
branch_12 = BatchNormalization(momentum=0.8)(branch_12)
branch_12 = Dense(128, kernel_initializer = "he_uniform")(branch_12)
branch_12 = LeakyReLU(0.2)(branch_12)
branch_12 = BatchNormalization(momentum=0.8)(branch_12)
#Output Layer12
branch_12_output = Dense(numerical, activation = "sigmoid")(branch_12)
#Combined output
combined_output = concatenate([branch_1_output, branch_2_output, branch_3_output,branch_4_output,branch_5_output,branch_6_output,branch_7_output,branch_8_output,branch_9_output,branch_10_output,branch_11_output,branch_12_output])
#Return model
return Model(inputs = noise, outputs = combined_output)
generator = define_generator(location_dummy.shape[1],country_dummy.shape[1],gender_dummy.shape[1],vis_wuhan_dummy.shape[1],from_wuhan_dummy.shape[1],symptom1_dummy.shape[1],symptom2_dummy.shape[1],symptom3_dummy.shape[1],symptom4_dummy.shape[1],symptom5_dummy.shape[1],symptom6_dummy.shape[1],numerical_df_rescaled.shape[1])
generator.summary()
定义鉴别器
鉴别器模型将从我们的数据(例如矢量)中获取样本,并输出关于样本是真实还是假的分类预测。这是一个二进制分类问题,因此在输出层中使用sigmoid 激活函数,在模型编译中使用二进制交叉熵损失函数。使用学习率LR为0.0002且建议的beta1动量值为0.5的Adam优化算法。
def define_discriminator(inputs_n):
#Input from generator
d_input = Input(shape = (inputs_n,))
d = Dense(128, kernel_initializer="he_uniform")(d_input)
d = LeakyReLU(0.2)(d)
d = Dense(64, kernel_initializer="he_uniform")(d)
d = LeakyReLU(0.2)(d)
d = Dense(32, kernel_initializer="he_uniform")(d)
d = LeakyReLU(0.2)(d)
d = Dense(16, kernel_initializer="he_uniform")(d)
d = LeakyReLU(0.2)(d)
d = Dense(8, kernel_initializer="he_uniform")(d)
d = LeakyReLU(0.2)(d)
#Output Layer
d_output = Dense(1, activation = "sigmoid")(d)
#compile and return model
model = Model(inputs = d_input, outputs = d_output)
model.compile(loss = "binary_crossentropy", optimizer = Adam(lr=0.0002, beta_1=0.5), metrics = ["accuracy"])
return model
inputs_n = location_dummy.shape[1]+country_dummy.shape[1]+gender_dummy.shape[1]+vis_wuhan_dummy.shape[1]+from_wuhan_dummy.shape[1]+symptom1_dummy.shape[1]+symptom2_dummy.shape[1]+symptom3_dummy.shape[1]+symptom4_dummy.shape[1]+symptom5_dummy.shape[1]+symptom6_dummy.shape[1]+numerical_df_rescaled.shape[1]
discriminator = define_discriminator(inputs_n)
discriminator.summary()
将生成器和鉴别器组合为GAN模型并完成训练。考虑了7,000个时期,并考虑了完整的少数派训练数据。
Def define_complete_gan(generator, discriminator):
discriminator.trainable = False
gan_output = discriminator(generator.output)
#Initialize gan
model = Model(inputs = generator.input, outputs = gan_output)
#Model Compilation
model.compile(loss = "binary_crossentropy", optimizer = Adam(lr=0.0002, beta_1=0.5))
return model
completegan = define_complete_gan(generator, discriminator)
def gan_train(gan, generator, discriminator, catsh1,catsh2,catsh3,catsh4,catsh5,catsh6,catsh7,catsh8,catsh9,catsh10,catsh11,numerical, latent_dim, n_epochs, n_batch, n_eval):
#Upddte Discriminator with half batch size
half_batch = int(n_batch / 2)
discriminator_loss = []
generator_loss = []
#generate class labels for fake and real
valid = np.ones((half_batch, 1))
y_gan = np.ones((n_batch, 1))
fake = np.zeros((half_batch, 1))
#training
for i in range(n_epochs):
#select random batch from real categorical and numerical data
idx = np.random.randint(0, catsh1.shape[0], half_batch)
location_real = catsh1[idx]
country_real = catsh2[idx]
gender_real = catsh3[idx]
vis_wuhan_real = catsh4[idx]
from_wuhan_real = catsh5[idx]
symptom1_real = catsh6[idx]
symptom2_real = catsh7[idx]
symptom3_real = catsh8[idx]
symptom4_real = catsh9[idx]
symptom5_real = catsh10[idx]
symptom6_real = catsh11[idx]
numerical_real = numerical_df_rescaled[idx]
#concatenate categorical and numerical data for the discriminator
real_data = np.concatenate([location_real, country_real, gender_real,vis_wuhan_real,from_wuhan_real,symptom1_real,symptom2_real,symptom3_real,symptom4_real,symptom5_real,symptom6_real,numerical_real], axis = 1)
#generate fake samples from the noise
noise = np.random.normal(0, 1, (half_batch, latent_dim))
fake_data = generator.predict(noise)
#train the discriminator and return losses and acc
d_loss_real, da_real = discriminator.train_on_batch(real_data, valid)
d_loss_fake, da_fake = discriminator.train_on_batch(fake_data, fake)
d_loss = 0.5 * np.add(d_loss_real, d_loss_fake)
discriminator_loss.append(d_loss)
#generate noise for generator input and train the generator (to have the discriminator label samples as valid)
noise = np.random.normal(0, 1, (n_batch, latent_dim))
g_loss = gan.train_on_batch(noise, y_gan)
generator_loss.append(g_loss)
#evaluate progress
if (i+1) % n_eval == 0:
print ("Epoch: %d [Discriminator loss: %f] [Generator loss: %f]" % (i + 1, d_loss, g_loss))
plt.figure(figsize = (20, 10))
plt.plot(generator_loss, label = "Generator loss")
plt.plot(discriminator_loss, label = "Discriminator loss")
plt.title("Stats from training GAN")
plt.grid()
plt.legend()
latent_dim = 100
gan_train(completegan, generator, discriminator, location_dummy.values,country_dummy.values,gender_dummy.values,vis_wuhan_dummy.values,from_wuhan_dummy.values,symptom1_dummy.values,symptom2_dummy.values,symptom3_dummy.values,symptom4_dummy.values,symptom5_dummy.values,symptom6_dummy.values,numerical_df_rescaled, latent_dim, n_epochs = 7000, n_batch = 63, n_eval = 200)

训练后的模型用于生成少数类的其他96条记录,以对每个类进行均等分割(159)。现在将生成的数值数据与原始数据的均值,标准差和方差进行比较;并根据每个类别的计数比较类别数据。
noise = np.random.normal(0, 1, (96, 100))
generated_mixed_data = generator.predict(noise)
columns=list(location_dummy.columns)+list(country_dummy.columns)+list(gender_dummy.columns)+list(vis_wuhan_dummy.columns)+list(from_wuhan_dummy.columns)+list(symptom1_dummy.columns)+list(symptom2_dummy.columns)+list(symptom3_dummy.columns)+list(symptom4_dummy.columns)+list(symptom5_dummy.columns)+list(symptom6_dummy.columns)+list(numerical_df.columns)
mixed_gen_df = pd.DataFrame(data = generated_mixed_data, columns = columns)
mixed_gen_df.iloc[:,:-3] = np.round(mixed_gen_df.iloc[:,:-3])
mixed_gen_df.iloc[:,-2:] = scaling.inverse_transform(mixed_gen_df.iloc[:,-2:])
#Original Data
original_df = pd.concat([location_dummy,country_dummy,gender_dummy,vis_wuhan_dummy,from_wuhan_dummy,symptom1_dummy,symptom2_dummy,symptom3_dummy,symptom4_dummy,symptom5_dummy,symptom6_dummy,numerical_df], axis = 1)
def normal_distribution(org, noise):
org_x = np.linspace(org.min(), org.max(), len(org))
noise_x = np.linspace(noise.min(), noise.max(), len(noise))
org_y = scipy.stats.norm.pdf(org_x, org.mean(), org.std())
noise_y = scipy.stats.norm.pdf(noise_x, noise.mean(), noise.std())
n, bins, patches = plt.hist([org, noise], density = True, alpha = 0.5, color = ["green", "red"])
xmin, xmax = plt.xlim()
plt.plot(org_x, org_y, color = "green", label = "Original data", alpha = 0.5)
plt.plot(noise_x, noise_y, color = "red", label = "Generated data", alpha = 0.5)
title = f"Original data mean {np.round(org.mean(), 4)}, Original data std {np.round(org.std(), 4)}, Original data var {np.round(org.var(), 4)}
Generated data mean {np.round(noise.mean(), 4)}, Generated data {np.round(noise.std(), 4)}, Generated data var {np.round(noise.var(), 2)}"
plt.title(title)
plt.legend()
plt.grid()
plt.show()
Numeric_columns=numerical_df.columns
for column in numerical_df.columns:
print(column, "Comparison between Original Data and Generated Data")
normal_distribution(original_df
, mixed_gen_df
)
原始数据和生成数据之间的年龄比较
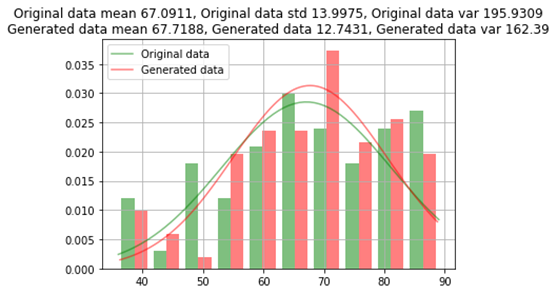
原始数据与生成的数据之间的比较
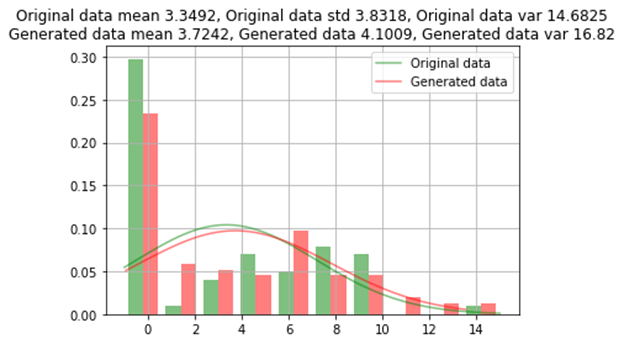
原始数据和生成的数据之间的类别比较
| 特征 | 原始数据 | 产生的数据 | ||
| 0 | 1 | 0 | 1 | |
| location_Hokkaido | 61 | 2 | 95 | 1 |
| gender_female | 49 | 14 | 60 | 36 |
| symptom2_ cough | 62 | 1 | 96 | 0 |
GAN过采样方法生成的数据几乎类似于原始数据,原始数据的误差约为1%。对于一些稀有类别,不会在所有类别值上生成数据。
遵循与原始研究中提到的相同的数据准备步骤,以查看通过使用GAN超采样与原始方法相比模型性能如何提高。所生成样本的独热编码数据被转换为原始数据帧格式。
# Getting Back Categorical Data in Original_Format from Dummies
location_filter_col = [col for col in mixed_gen_df if col.startswith('location')]
location=mixed_gen_df[location_filter_col]
location= pd.get_dummies(location).idxmax(1)
location= location.replace('location_', '', regex=True)
df_generated_data = pd.DataFrame()
df_generated_data['location']=location
country_filter_col = [col for col in mixed_gen_df if col.startswith('country')]
country=mixed_gen_df[country_filter_col]
country= pd.get_dummies(country).idxmax(1)
country= country.replace('country_', '', regex=True)
df_generated_data['country']=country
gender_filter_col = [col for col in mixed_gen_df if col.startswith('gender')]
gender=mixed_gen_df[gender_filter_col]
gender= pd.get_dummies(gender).idxmax(1)
gender= gender.replace('gender_', '', regex=True)
df_generated_data['gender']=gender
vis_wuhan_filter_col = [col for col in mixed_gen_df if col.startswith('vis_wuhan')]
vis_wuhan=mixed_gen_df[vis_wuhan_filter_col]
vis_wuhan= pd.get_dummies(vis_wuhan).idxmax(1)
vis_wuhan= vis_wuhan.replace('vis_wuhan_', '', regex=True)
df_generated_data['vis_wuhan']=vis_wuhan
from_wuhan_filter_col = [col for col in mixed_gen_df if col.startswith('from_wuhan')]
from_wuhan=mixed_gen_df[from_wuhan_filter_col]
from_wuhan= pd.get_dummies(from_wuhan).idxmax(1)
from_wuhan= from_wuhan.replace('from_wuhan_', '', regex=True)
df_generated_data['from_wuhan']=from_wuhan
symptom1_filter_col = [col for col in mixed_gen_df if col.startswith('symptom1')]
symptom1=mixed_gen_df[symptom1_filter_col]
symptom1= pd.get_dummies(symptom1).idxmax(1)
symptom1= symptom1.replace('symptom1_', '', regex=True)
df_generated_data['symptom1']=symptom1
symptom2_filter_col = [col for col in mixed_gen_df if col.startswith('symptom2')]
symptom2=mixed_gen_df[symptom2_filter_col]
symptom2= pd.get_dummies(symptom2).idxmax(1)
symptom2= symptom2.replace('symptom2_', '', regex=True)
df_generated_data['symptom2']=symptom2
symptom3_filter_col = [col for col in mixed_gen_df if col.startswith('symptom3')]
symptom3=mixed_gen_df[symptom3_filter_col]
symptom3= pd.get_dummies(symptom3).idxmax(1)
symptom3= symptom3.replace('symptom3_', '', regex=True)
df_generated_data['symptom3']=symptom3
symptom4_filter_col = [col for col in mixed_gen_df if col.startswith('symptom4')]
symptom4=mixed_gen_df[symptom4_filter_col]
symptom4= pd.get_dummies(symptom4).idxmax(1)
symptom4= symptom4.replace('symptom4_', '', regex=True)
df_generated_data['symptom4']=symptom4
symptom5_filter_col = [col for col in mixed_gen_df if col.startswith('symptom5')]
symptom5=mixed_gen_df[symptom5_filter_col]
symptom5= pd.get_dummies(symptom5).idxmax(1)
symptom5= symptom5.replace('symptom5_', '', regex=True)
df_generated_data['symptom5']=symptom5
symptom6_filter_col = [col for col in mixed_gen_df if col.startswith('symptom6')]
symptom6=mixed_gen_df[symptom6_filter_col]
symptom6= pd.get_dummies(symptom6).idxmax(1)
symptom6= symptom6.replace('symptom6_', '', regex=True)
df_generated_data['symptom6']=symptom6
df_generated_data['death']=1
df_generated_data['death']=1
df_generated_data[['age','diff_sym_hos']]=mixed_gen_df[['age','diff_sym_hos']]
df_generated_data = df_generated_data.fillna(np.nan,axis=0)
#Encoding Data
encoder_location = preprocessing.LabelEncoder()
encoder_country = preprocessing.LabelEncoder()
encoder_gender = preprocessing.LabelEncoder()
encoder_symptom1 = preprocessing.LabelEncoder()
encoder_symptom2 = preprocessing.LabelEncoder()
encoder_symptom3 = preprocessing.LabelEncoder()
encoder_symptom4 = preprocessing.LabelEncoder()
encoder_symptom5 = preprocessing.LabelEncoder()
encoder_symptom6 = preprocessing.LabelEncoder()
# Loading and Preparing Data
df = pd.read_csv('Covid_Train_Oct32020.csv')
df = df.drop('id',axis=1)
df = df.fillna(np.nan,axis=0)
df['age'] = df['age'].fillna(value=tdata['age'].mean())
df['sym_on'] = pd.to_datetime(df['sym_on'])
df['hosp_vis'] = pd.to_datetime(df['hosp_vis'])
df['sym_on']= df['sym_on'].map(dt.datetime.toordinal)
df['hosp_vis']= df['hosp_vis'].map(dt.datetime.toordinal)
df['diff_sym_hos']= df['hosp_vis'] - df['sym_on']
df = df.drop(['sym_on','hosp_vis'],axis=1)
df['location'] = encoder_location.fit_transform(df['location'].astype(str))
df['country'] = encoder_country.fit_transform(df['country'].astype(str))
df['gender'] = encoder_gender.fit_transform(df['gender'].astype(str))
df[['symptom1']] = encoder_symptom1.fit_transform(df['symptom1'].astype(str))
df[['symptom2']] = encoder_symptom2.fit_transform(df['symptom2'].astype(str))
df[['symptom3']] = encoder_symptom3.fit_transform(df['symptom3'].astype(str))
df[['symptom4']] = encoder_symptom4.fit_transform(df['symptom4'].astype(str))
df[['symptom5']] = encoder_symptom5.fit_transform(df['symptom5'].astype(str))
df[['symptom6']] = encoder_symptom6.fit_transform(df['symptom6'].astype(str))
# Encoding Generated Data
df_generated_data['location'] = encoder_location.transform(df_generated_data['location'].astype(str))
df_generated_data['country'] = encoder_country.transform(df_generated_data['country'].astype(str))
df_generated_data['gender'] = encoder_gender.transform(df_generated_data['gender'].astype(str))
df_generated_data[['symptom1']] = encoder_symptom1.transform(df_generated_data['symptom1'].astype(str))
df_generated_data[['symptom2']] = encoder_symptom2.transform(df_generated_data['symptom2'].astype(str))
df_generated_data[['symptom3']] = encoder_symptom3.transform(df_generated_data['symptom3'].astype(str))
df_generated_data[['symptom4']] = encoder_symptom4.transform(df_generated_data['symptom4'].astype(str))
df_generated_data[['symptom5']] = encoder_symptom5.transform(df_generated_data['symptom5'].astype(str))
df_generated_data[['symptom6']] = encoder_symptom6.transform(df_generated_data['symptom6'].astype(str))
df_generated_data[['diff_sym_hos']] = df_generated_data['diff_sym_hos'].astype(int)
模型比较
将原始数据分为训练和测试后,将GAN生成的数据添加到训练数据中,以将性能与基本模型进行比较。在实际(原始)分割测试数据上测试模型性能。
from sklearn.metrics import recall_score as rs
from sklearn.metrics import precision_score as ps
from sklearn.metrics import f1_score as fs
from sklearn.metrics import balanced_accuracy_score as bas
from sklearn.metrics import confusion_matrix as cm
import numpy as np
import pandas as pd
import datetime as dt
import sklearn
from scipy import stats
from sklearn import preprocessing
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import recall_score as rs
from sklearn.metrics import precision_score as ps
from sklearn.metrics import f1_score as fs
from sklearn.metrics import log_loss
rf = RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=2, max_features='auto',
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=2, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=100,
n_jobs=None, oob_score=False, random_state=None,
verbose=0, warm_start=False)
classifier = AdaBoostClassifier(rf,50,0.01,'SAMME.R',10)
#Seperate TV in Generated Data
X1 = df_generated_data.loc[:, df_generated_data.columns != 'death']
Y1 = df_generated_data['death']
#Seperate TV in Original Data
X = df.loc[:, df.columns != 'death']
Y = df['death']
#Splitting Original Data
X_train, X_test, Y_train, Y_test = train_test_split(X,Y,test_size=0.2,random_state=0)
#Appending Generated Data to X_train
X_train1=X_train.append(X1, sort=False)
Y_train1=Y_train.append(Y1)
classifier.fit(X_train1,np.array(Y_train1).reshape(Y_train1.shape[0],1))
pred = np.array(classifier.predict(X_test))
recall = rs(Y_test,pred)
precision = ps(Y_test,pred)
r1 = fs(Y_test,pred)
ma = classifier.score(X_test,Y_test)
print('*** Evaluation metrics for test dataset ***
')
print('Recall Score: ',recall)
print('Precision Score: ',precision)
print('F1 Score: ',f1)
print('Accuracy: ',ma)
| 公制 | 基本模型得分* | 用增强的生成数据评分 |
| 召回分数 | 0.75 | 0.83 |
| 精度分数 | 1 | 1 |
| F1分数 | 0.86 | 0.9 |
| 准确性 | 0.9 | 0.95 |
资料来源:表3基本模型指标
结论
与基本模型相比,所提出的模型提供了更加准确和可靠的结果,表明基于GAN的过采样克服了不平衡数据的局限性,并适当地扩充了少数类。
欢迎关注磐创AI博客站:
http://panchuang.net/
sklearn机器学习中文官方文档:
http://sklearn123.com/
欢迎关注磐创博客资源汇总站:
http://docs.panchuang.net/