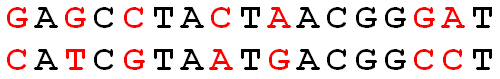Problem
Given two strings ss and tt of equal length, the Hamming distance between ss and tt, denoted dH(s,t)dH(s,t), is the number of corresponding symbols that differ in ss and tt. See Figure 2.
Given: Two DNA strings ss and tt of equal length (not exceeding 1 kbp).
Return: The Hamming distance dH(s,t)dH(s,t).
Sample Dataset
GAGCCTACTAACGGGAT CATCGTAATGACGGCCT
Sample Output
7
方法一:
s = 'GAGCCTACTAACGGGAT'
t = 'CATCGTAATGACGGCCT'
count = 0
for i in xrange(len(s)):
if s[i] != t[i]:
count +=1
print count
方法二:
### Counting Point Mutations ###
fh = open('/Users/DONG/Downloads/rosalind_hamm.txt', 'rt')
seq = fh.readlines()
a,b = seq[0].strip(),seq[1].strip()
hammingDistance = 0
for i in range(len(a)):
if a[i] != b[i]:
hammingDistance += 1
print (hammingDistance)