
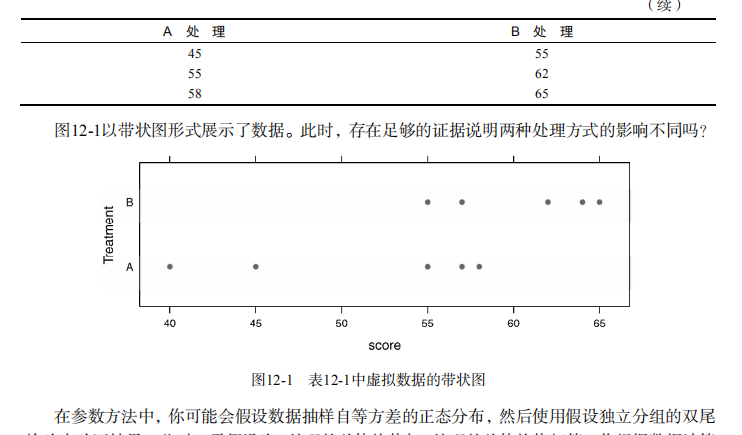





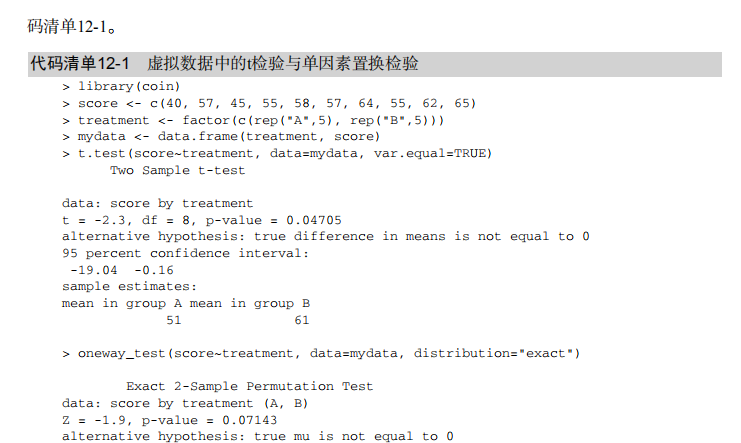







#-------------------------------------------------------------------------# # R in Action (2nd ed): Chapter 12 # # Resampling statistics and bootstrapping # # requires packages coin, multcomp, vcd, MASS, lmPerm, boot # # install.packages(c("coin","multcomp", "vcd", "MASS", "boot")) # # Follow chapter instructions for installing lmPerm # #-------------------------------------------------------------------------# par(ask=TRUE) # Listing 12.1 - t-test vs. oneway permutation test for the hypothetical data library(coin) score <- c(40, 57, 45, 55, 58, 57, 64, 55, 62, 65) treatment <- factor(c(rep("A",5), rep("B",5))) mydata <- data.frame(treatment, score) t.test(score~treatment, data=mydata, var.equal=TRUE) oneway_test(score~treatment, data=mydata, distribution="exact") # Wilcoxon Mann-Whitney U test UScrime <- transform(UScrime, So = factor(So)) wilcox_test(Prob ~ So, data=UScrime, distribution="exact") # k sample test library(multcomp) set.seed(1234) # make results reproducible oneway_test(response~trt, data=cholesterol, distribution=approximate(B=9999)) # independence in contingency tables library(coin) library(vcd) Arthritis <- transform(Arthritis, Improved = as.factor(as.numeric(Improved))) set.seed(1234) chisq_test(Treatment~Improved, data=Arthritis, distribution=approximate(B=9999)) # independence between numeric variables states <- as.data.frame(state.x77) set.seed(1234) spearman_test(Illiteracy ~ Murder, data=states, distribution=approximate(B=9999)) # dependent 2-sample and k-sample tests library(coin) library(MASS) wilcoxsign_test(U1 ~ U2, data=UScrime, distribution="exact") # Listing 12.2 - Permutation tests for simple linear regression library(lmPerm) set.seed(1234) fit <- lmp(weight ~ height, data=women, perm="Prob") summary(fit) # Listing 12.3 - Permutation tests for polynomial regression library(lmPerm) set.seed(1234) fit <- lmp(weight ~ height + I(height^2), data=women, perm="Prob") summary(fit) # Listing 12.4 - Permutation tests for multiple regression library(lmPerm) set.seed(1234) states <- as.data.frame(state.x77) fit <- lmp(Murder ~ Population + Illiteracy+Income+Frost,data=states, perm="Prob") summary(fit) # Listing 12.5 - Permutation test for One-Way ANOVA library(lmPerm) library(multcomp) set.seed(1234) fit <- lmp(response ~ trt, data=cholesterol, perm="Prob") anova(fit) # Listing 12.6 - Permutation test for One-Way ANCOVA library(lmPerm) set.seed(1234) fit <- lmp(weight ~ gesttime + dose, data=litter, perm="Prob") anova(fit) # Listing 12.7 - Permutation test for Two-way ANOVA library(lmPerm) set.seed(1234) fit <- lmp(len ~ supp*dose, data=ToothGrowth, perm="Prob") anova(fit) # bootstrapping a single statistic (R2) rsq <- function(formula, data, indices) { d <- data[indices,] fit <- lm(formula, data=d) return(summary(fit)$r.square) } library(boot) set.seed(1234) results <- boot(data=mtcars, statistic=rsq, R=1000, formula=mpg~wt+disp) print(results) plot(results) boot.ci(results, type=c("perc", "bca")) # bootstrapping several statistics (regression coefficients) bs <- function(formula, data, indices) { d <- data[indices,] fit <- lm(formula, data=d) return(coef(fit)) } library(boot) set.seed(1234) results <- boot(data=mtcars, statistic=bs, R=1000, formula=mpg~wt+disp) print(results) plot(results, index=2) boot.ci(results, type="bca", index=2) boot.ci(results, type="bca", index=3)