

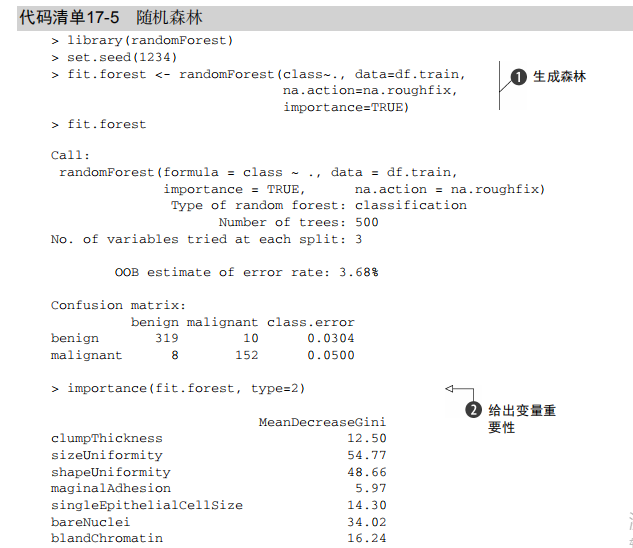



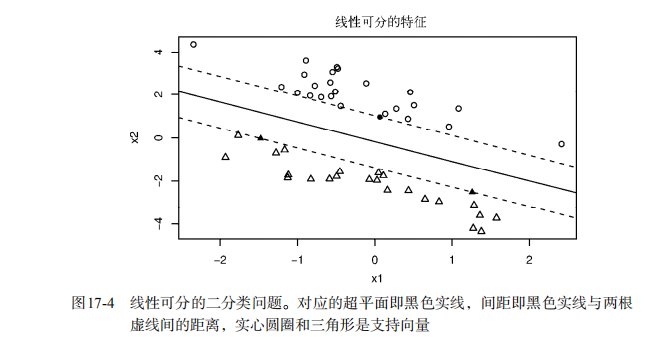

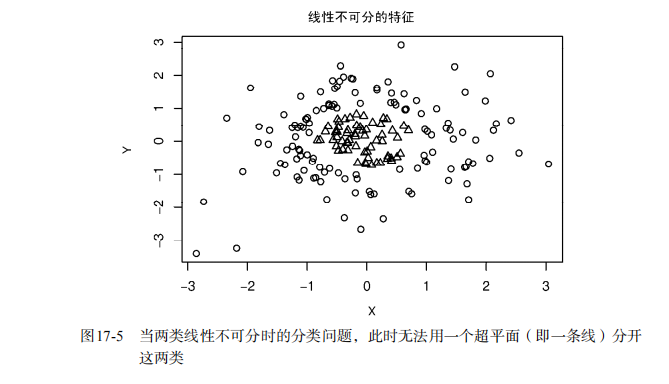

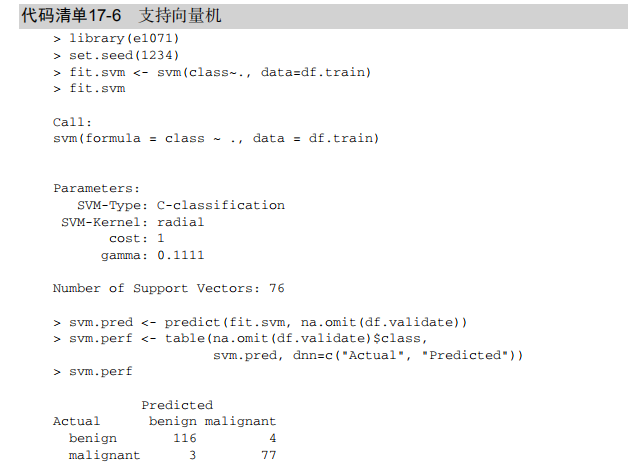


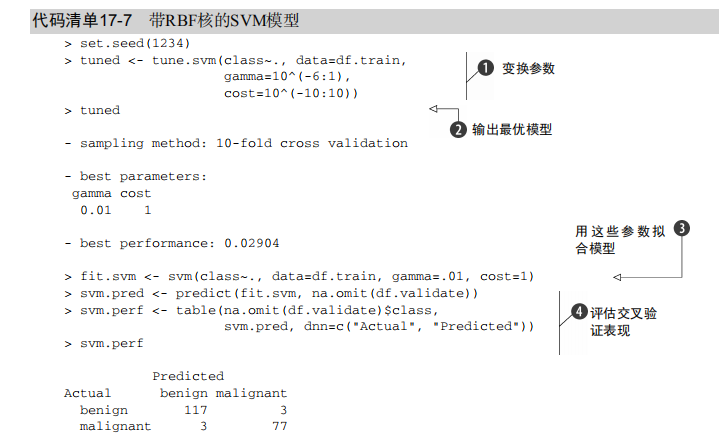







#-----------------------------------------------------------------------------# # R in Action (2nd ed): Chapter 17 # # Classification # # requires packaged rpart, party, randomForest, kernlab, rattle # # install.packages(c("rpart", "party", "randomForest", "e1071", "rpart.plot") # # install.packages(rattle, dependencies = c("Depends", "Suggests")) # #-----------------------------------------------------------------------------# par(ask=TRUE) # Listing 17.1 - Prepare the breast cancer data loc <- "http://archive.ics.uci.edu/ml/machine-learning-databases/" ds <- "breast-cancer-wisconsin/breast-cancer-wisconsin.data" url <- paste(loc, ds, sep="") breast <- read.table(url, sep=",", header=FALSE, na.strings="?") names(breast) <- c("ID", "clumpThickness", "sizeUniformity", "shapeUniformity", "maginalAdhesion", "singleEpithelialCellSize", "bareNuclei", "blandChromatin", "normalNucleoli", "mitosis", "class") df <- breast[-1] df$class <- factor(df$class, levels=c(2,4), labels=c("benign", "malignant")) set.seed(1234) train <- sample(nrow(df), 0.7*nrow(df)) df.train <- df[train,] df.validate <- df[-train,] table(df.train$class) table(df.validate$class) # Listing 17.2 - Logistic regression with glm() fit.logit <- glm(class~., data=df.train, family=binomial()) summary(fit.logit) prob <- predict(fit.logit, df.validate, type="response") logit.pred <- factor(prob > .5, levels=c(FALSE, TRUE), labels=c("benign", "malignant")) logit.perf <- table(df.validate$class, logit.pred, dnn=c("Actual", "Predicted")) logit.perf # Listing 17.3 - Creating a classical decision tree with rpart() library(rpart) set.seed(1234) dtree <- rpart(class ~ ., data=df.train, method="class", parms=list(split="information")) dtree$cptable plotcp(dtree) dtree.pruned <- prune(dtree, cp=.0125) library(rpart.plot) prp(dtree.pruned, type = 2, extra = 104, fallen.leaves = TRUE, main="Decision Tree") dtree.pred <- predict(dtree.pruned, df.validate, type="class") dtree.perf <- table(df.validate$class, dtree.pred, dnn=c("Actual", "Predicted")) dtree.perf # Listing 17.4 - Creating a conditional inference tree with ctree() library(party) fit.ctree <- ctree(class~., data=df.train) plot(fit.ctree, main="Conditional Inference Tree") ctree.pred <- predict(fit.ctree, df.validate, type="response") ctree.perf <- table(df.validate$class, ctree.pred, dnn=c("Actual", "Predicted")) ctree.perf # Listing 17.5 - Random forest library(randomForest) set.seed(1234) fit.forest <- randomForest(class~., data=df.train, na.action=na.roughfix, importance=TRUE) fit.forest importance(fit.forest, type=2) forest.pred <- predict(fit.forest, df.validate) forest.perf <- table(df.validate$class, forest.pred, dnn=c("Actual", "Predicted")) forest.perf # Listing 17.6 - A support vector machine library(e1071) set.seed(1234) fit.svm <- svm(class~., data=df.train) fit.svm svm.pred <- predict(fit.svm, na.omit(df.validate)) svm.perf <- table(na.omit(df.validate)$class, svm.pred, dnn=c("Actual", "Predicted")) svm.perf # Listing 17.7 Tuning an RBF support vector machine (this can take a while) set.seed(1234) tuned <- tune.svm(class~., data=df.train, gamma=10^(-6:1), cost=10^(-10:10)) tuned fit.svm <- svm(class~., data=df.train, gamma=.01, cost=1) svm.pred <- predict(fit.svm, na.omit(df.validate)) svm.perf <- table(na.omit(df.validate)$class, svm.pred, dnn=c("Actual", "Predicted")) svm.perf # Listing 17.8 Function for assessing binary classification accuracy performance <- function(table, n=2){ if(!all(dim(table) == c(2,2))) stop("Must be a 2 x 2 table") tn = table[1,1] fp = table[1,2] fn = table[2,1] tp = table[2,2] sensitivity = tp/(tp+fn) specificity = tn/(tn+fp) ppp = tp/(tp+fp) npp = tn/(tn+fn) hitrate = (tp+tn)/(tp+tn+fp+fn) result <- paste("Sensitivity = ", round(sensitivity, n) , " Specificity = ", round(specificity, n), " Positive Predictive Value = ", round(ppp, n), " Negative Predictive Value = ", round(npp, n), " Accuracy = ", round(hitrate, n), " ", sep="") cat(result) } # Listing 17.9 - Performance of breast cancer data classifiers performance(dtree.perf) performance(ctree.perf) performance(forest.perf) performance(svm.perf) # Using Rattle Package for data mining loc <- "http://archive.ics.uci.edu/ml/machine-learning-databases/" ds <- "pima-indians-diabetes/pima-indians-diabetes.data" url <- paste(loc, ds, sep="") diabetes <- read.table(url, sep=",", header=FALSE) names(diabetes) <- c("npregant", "plasma", "bp", "triceps", "insulin", "bmi", "pedigree", "age", "class") diabetes$class <- factor(diabetes$class, levels=c(0,1), labels=c("normal", "diabetic")) library(rattle) rattle()