# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load in
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns # data visualization library
import matplotlib.pyplot as plt
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import time
from subprocess import check_output
data = pd.read_csv('../input/data.csv')
data.head() # head method show only first 5 rows
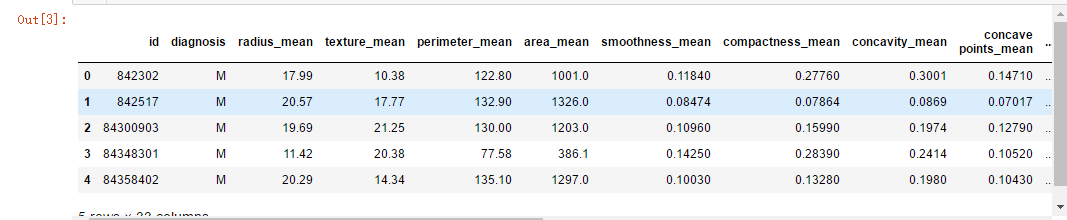
# feature names as a list
col = data.columns # .columns gives columns names in data
print(col)

# y includes our labels and x includes our features
y = data.diagnosis # M or B
list = ['Unnamed: 32','id','diagnosis']
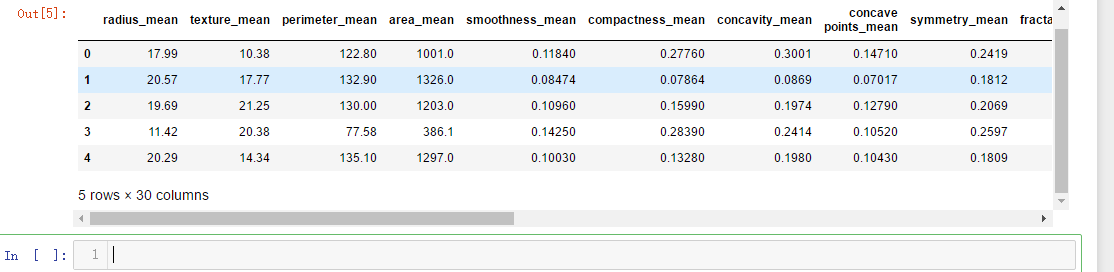
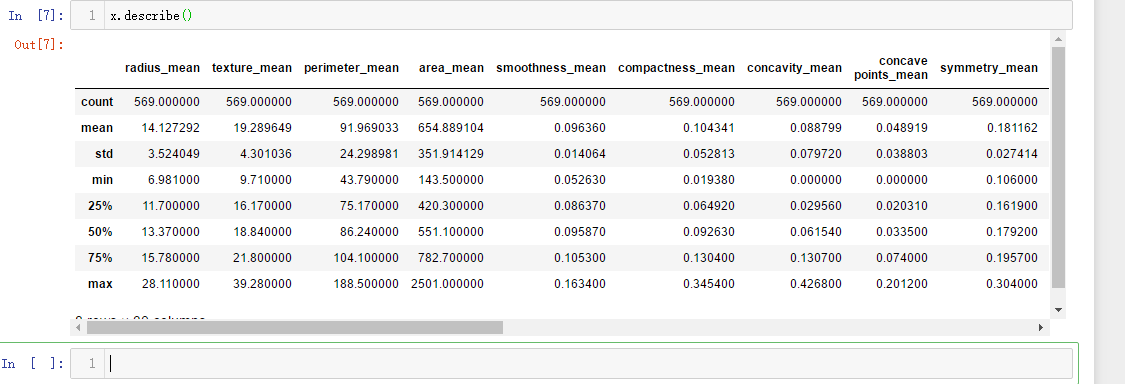
x = data.drop(list,axis = 1 )
x.head()
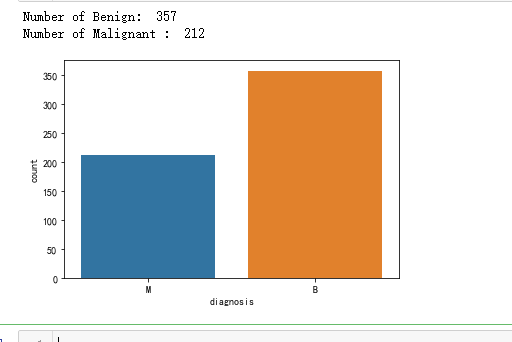
ax = sns.countplot(y,label="Count") # M = 212, B = 357
B, M = y.value_counts()
print('Number of Benign: ',B)
print('Number of Malignant : ',M)
# first ten features
data_dia = y
data = x
data_n_2 = (data - data.mean()) / (data.std()) # standardization
data = pd.concat([y,data_n_2.iloc[:,0:10]],axis=1)
data = pd.melt(data,id_vars="diagnosis",
var_name="features",
value_name='value')
plt.figure(figsize=(10,10))
sns.violinplot(x="features", y="value", hue="diagnosis", data=data,split=True, inner="quart")
plt.xticks(rotation=90)
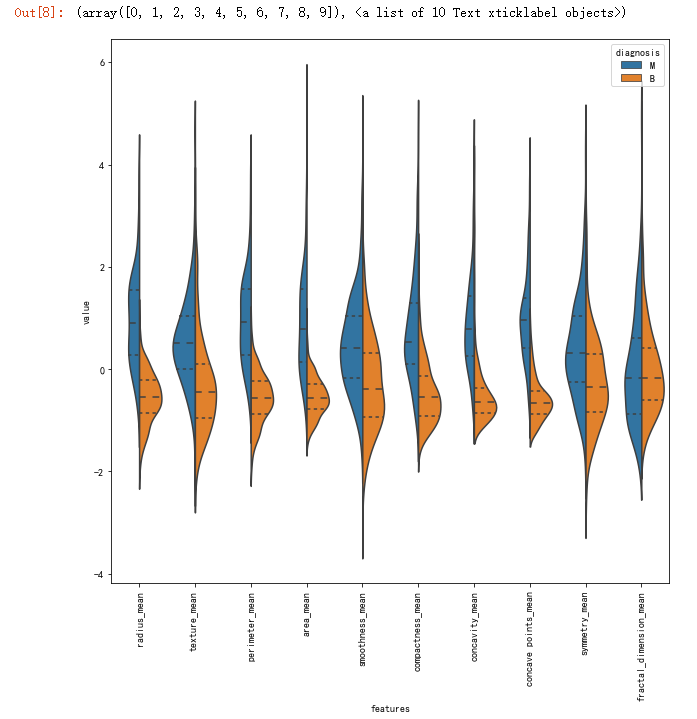
# Second ten features
data = pd.concat([y,data_n_2.iloc[:,10:20]],axis=1)
data = pd.melt(data,id_vars="diagnosis",
var_name="features",
value_name='value')
plt.figure(figsize=(10,10))
sns.violinplot(x="features", y="value", hue="diagnosis", data=data,split=True, inner="quart")
plt.xticks(rotation=90)
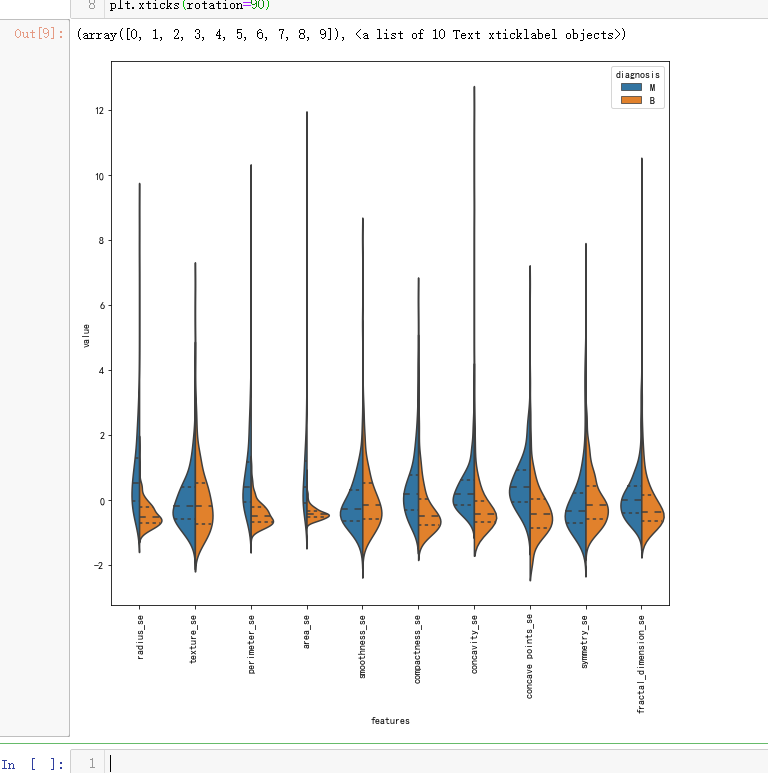
# Second ten features
data = pd.concat([y,data_n_2.iloc[:,20:31]],axis=1)
data = pd.melt(data,id_vars="diagnosis",
var_name="features",
value_name='value')
plt.figure(figsize=(10,10))
sns.violinplot(x="features", y="value", hue="diagnosis", data=data,split=True, inner="quart")
plt.xticks(rotation=90)
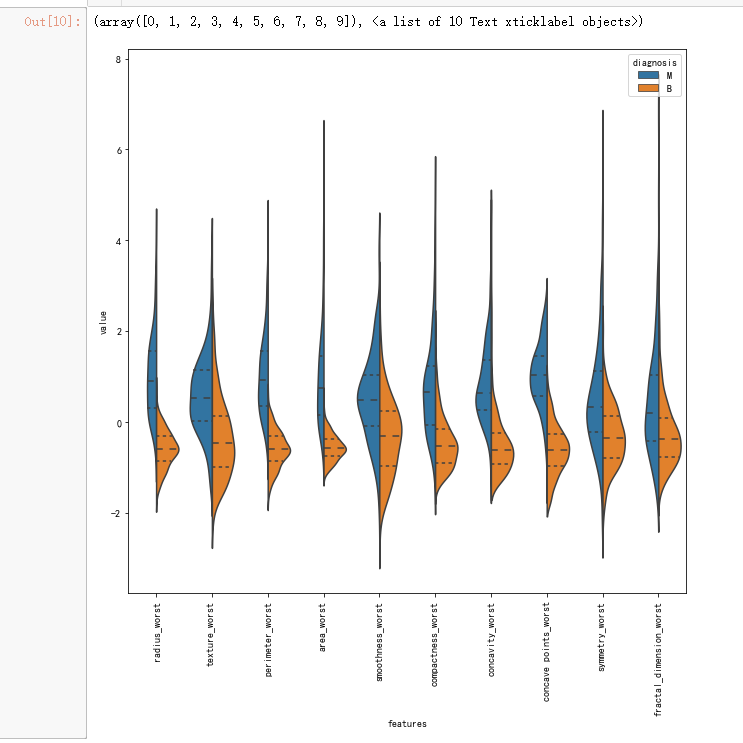
# As an alternative of violin plot, box plot can be used
# box plots are also useful in terms of seeing outliers
# I do not visualize all features with box plot
# In order to show you lets have an example of box plot
# If you want, you can visualize other features as well.
plt.figure(figsize=(10,10))
sns.boxplot(x="features", y="value", hue="diagnosis", data=data)
plt.xticks(rotation=90)
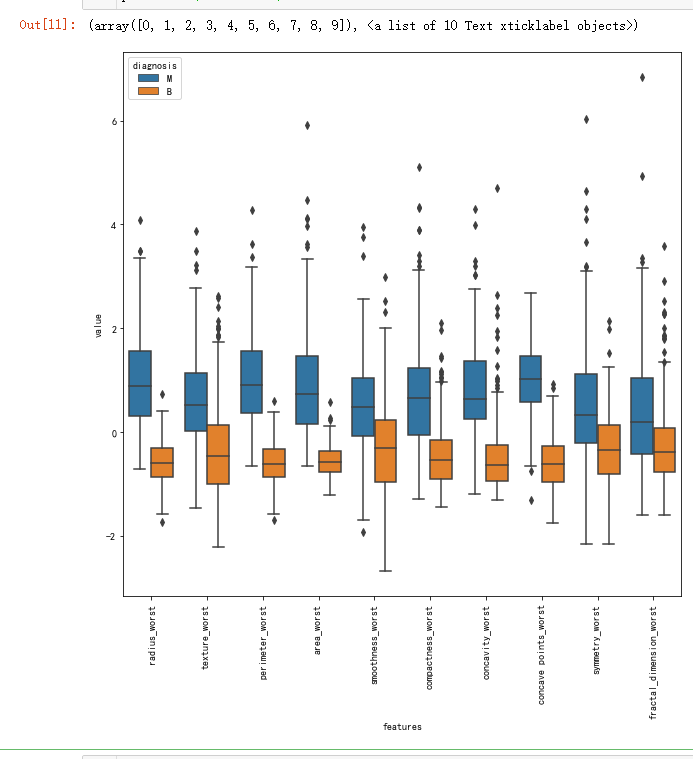
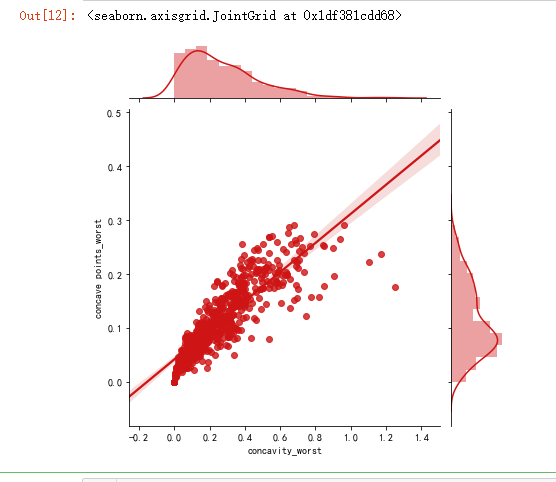
sns.jointplot(x.loc[:,'concavity_worst'], x.loc[:,'concave points_worst'], kind="regg", color="#ce1414")
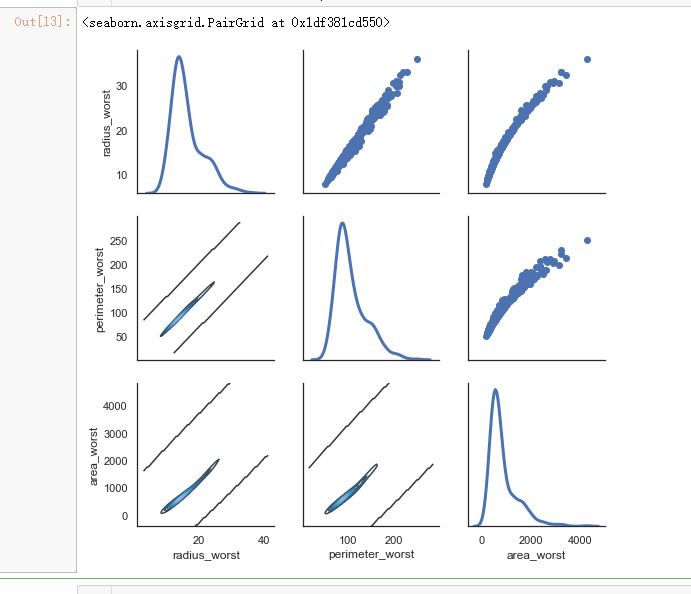
sns.set(style="white")
df = x.loc[:,['radius_worst','perimeter_worst','area_worst']]
g = sns.PairGrid(df, diag_sharey=False)
g.map_lower(sns.kdeplot, cmap="Blues_d")
g.map_upper(plt.scatter)
g.map_diag(sns.kdeplot, lw=3)
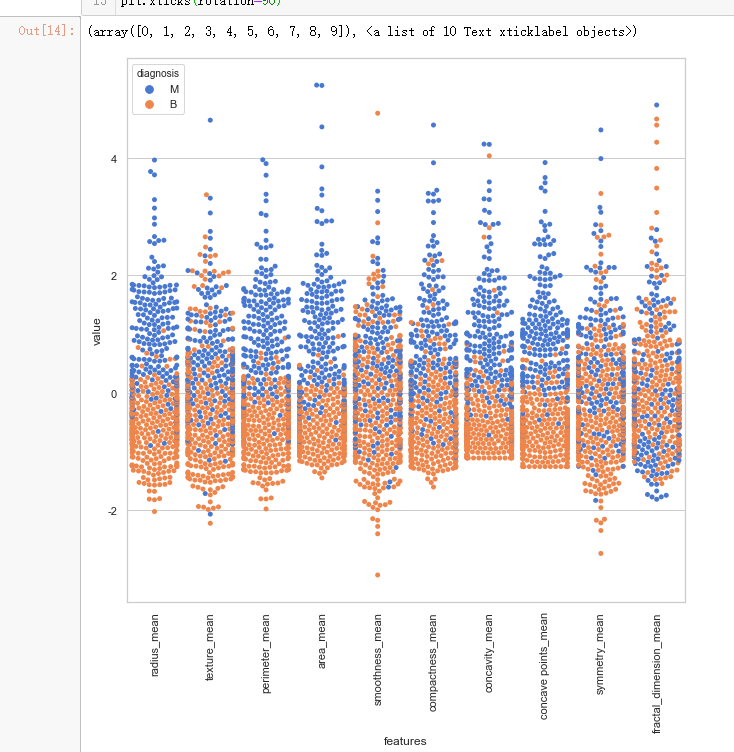
sns.set(style="whitegrid", palette="muted")
data_dia = y
data = x
data_n_2 = (data - data.mean()) / (data.std()) # standardization
data = pd.concat([y,data_n_2.iloc[:,0:10]],axis=1)
data = pd.melt(data,id_vars="diagnosis",
var_name="features",
value_name='value')
plt.figure(figsize=(10,10))
tic = time.time()
sns.swarmplot(x="features", y="value", hue="diagnosis", data=data)
plt.xticks(rotation=90)
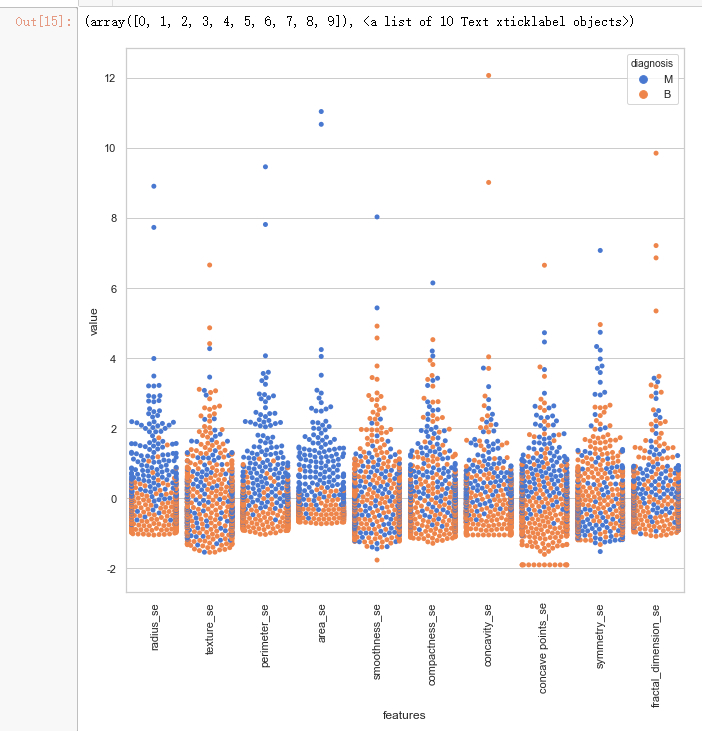
data = pd.concat([y,data_n_2.iloc[:,10:20]],axis=1)
data = pd.melt(data,id_vars="diagnosis",
var_name="features",
value_name='value')
plt.figure(figsize=(10,10))
sns.swarmplot(x="features", y="value", hue="diagnosis", data=data)
plt.xticks(rotation=90)
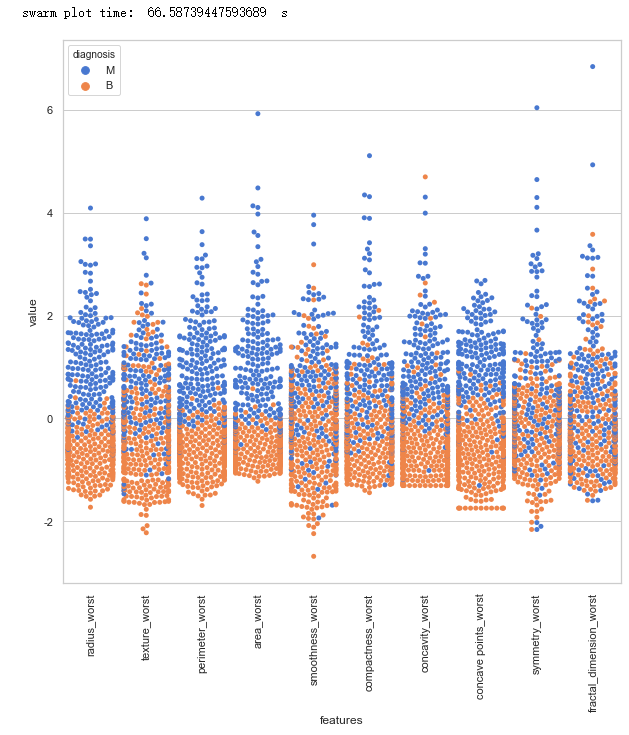
data = pd.concat([y,data_n_2.iloc[:,20:31]],axis=1)
data = pd.melt(data,id_vars="diagnosis",
var_name="features",
value_name='value')
plt.figure(figsize=(10,10))
sns.swarmplot(x="features", y="value", hue="diagnosis", data=data)
toc = time.time()
plt.xticks(rotation=90)
print("swarm plot time: ", toc-tic ," s")
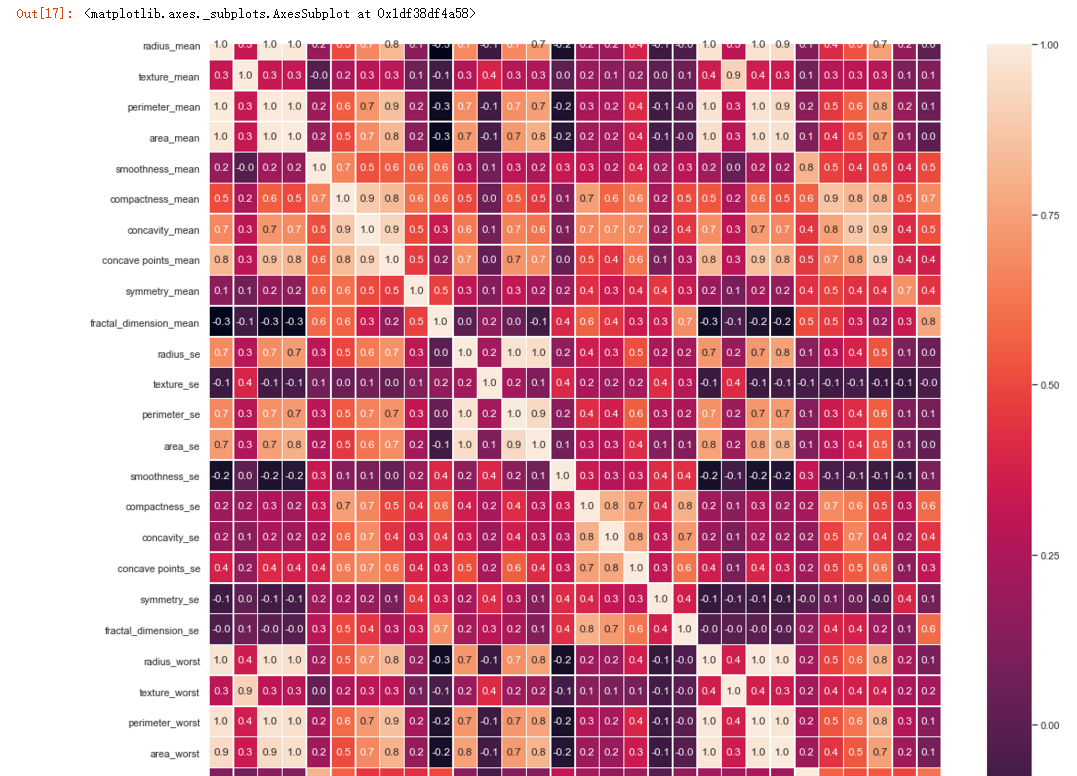
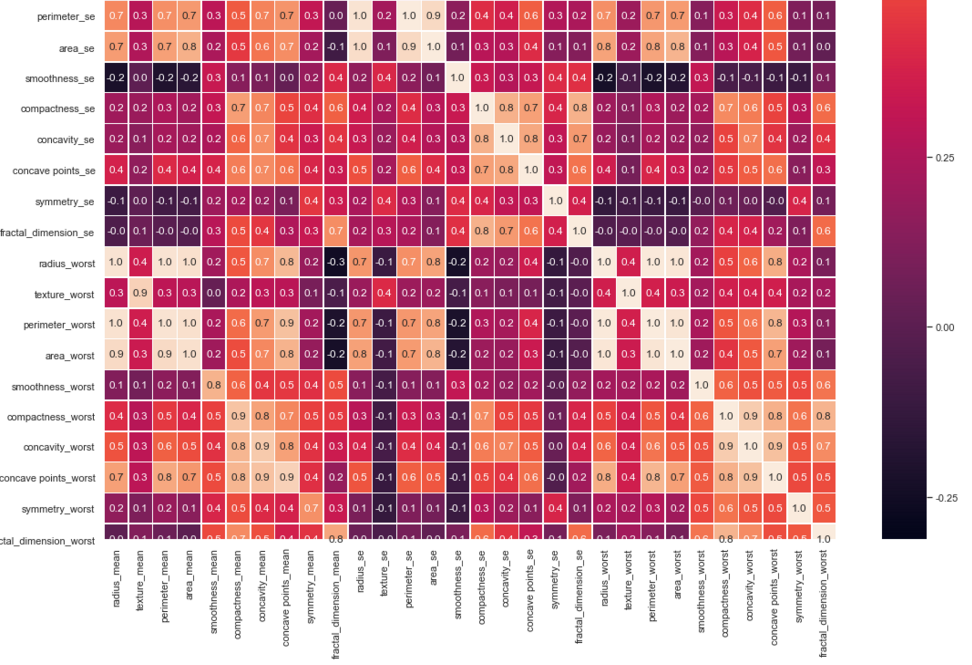
#correlation map
f,ax = plt.subplots(figsize=(18, 18))
sns.heatmap(x.corr(), annot=True, linewidths=.5, fmt= '.1f',ax=ax)