# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load in
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import os, sys
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import skimage.io
from skimage.transform import resize
#from imgaug import augmenters as iaa
from tqdm import tqdm
import PIL
from PIL import Image, ImageOps
import cv2
from sklearn.utils import class_weight, shuffle
from keras.losses import binary_crossentropy
from keras.applications.resnet50 import preprocess_input
import keras.backend as K
import tensorflow as tf
from sklearn.metrics import f1_score, fbeta_score
from keras.utils import Sequence
from keras.utils import to_categorical
from sklearn.model_selection import train_test_split
WORKERS = 2
CHANNEL = 3
import warnings
warnings.filterwarnings("ignore")
IMG_SIZE = 512
NUM_CLASSES = 5
SEED = 77
TRAIN_NUM = 1000 # use 1000 when you just want to explore new idea, use -1 for full train
df_train = pd.read_csv('F:\kaggleDataSet\diabeticRetinopathy\trainLabels19.csv')
df_test = pd.read_csv('F:\kaggleDataSet\diabeticRetinopathy\testImages19.csv')
x = df_train['id_code']
y = df_train['diagnosis']
x, y = shuffle(x, y, random_state=SEED)
train_x, valid_x, train_y, valid_y = train_test_split(x, y, test_size=0.15,stratify=y, random_state=SEED)
print(train_x.shape, train_y.shape, valid_x.shape, valid_y.shape)
train_y.hist()
valid_y.hist()

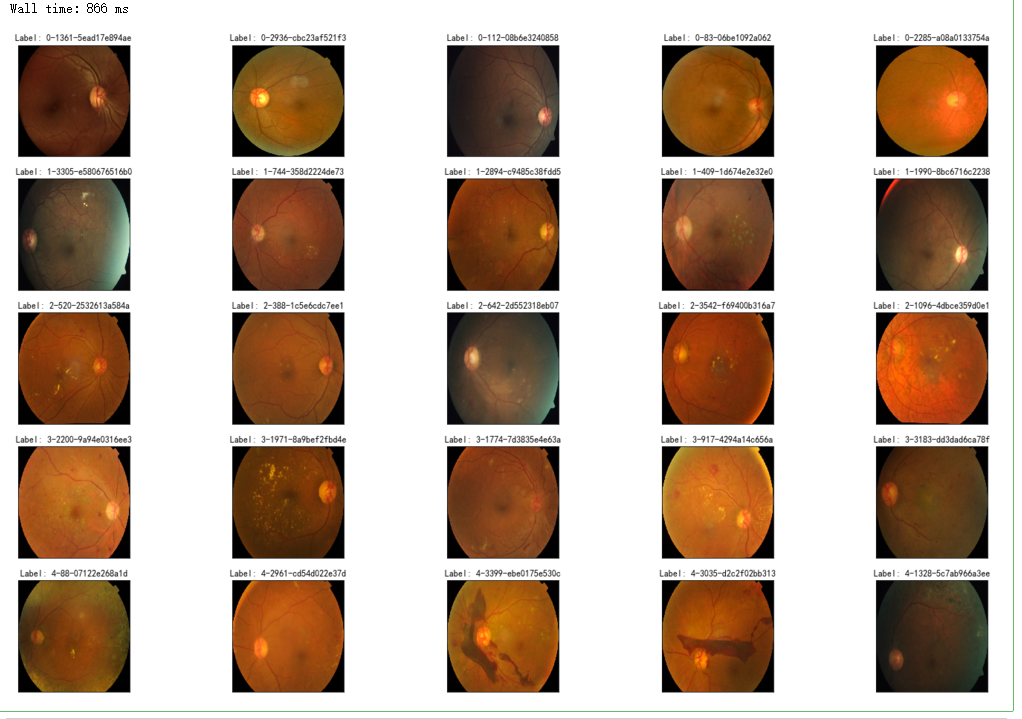
%%time
fig = plt.figure(figsize=(25, 16))
# display 10 images from each class
for class_id in sorted(train_y.unique()):
for i, (idx, row) in enumerate(df_train.loc[df_train['diagnosis'] == class_id].sample(5, random_state=SEED).iterrows()):
ax = fig.add_subplot(5, 5, class_id * 5 + i + 1, xticks=[], yticks=[])
path="F:\kaggleDataSet\diabeticRetinopathy\resized train 19\"+str(row['id_code'])+".jpg"
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.resize(image, (IMG_SIZE, IMG_SIZE))
plt.imshow(image)
ax.set_title('Label: %d-%d-%s' % (class_id, idx, row['id_code']) )
%%time
fig = plt.figure(figsize=(25, 16))
for class_id in sorted(train_y.unique()):
for i, (idx, row) in enumerate(df_train.loc[df_train['diagnosis'] == class_id].sample(5, random_state=SEED).iterrows()):
ax = fig.add_subplot(5, 5, class_id * 5 + i + 1, xticks=[], yticks=[])
path="F:\kaggleDataSet\diabeticRetinopathy\resized train 19\"+str(row['id_code'])+".jpg"
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
# image=cv2.addWeighted ( image, 0 , cv2.GaussianBlur( image , (0 ,0 ) , 10) ,-4 ,128)
image = cv2.resize(image, (IMG_SIZE, IMG_SIZE))
plt.imshow(image, cmap='gray')
ax.set_title('Label: %d-%d-%s' % (class_id, idx, row['id_code']) )

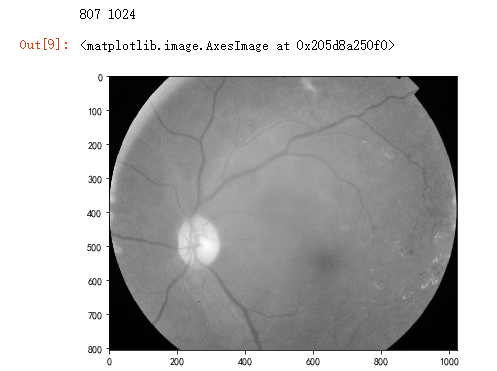
dpi = 80 #inch
# path=f"../input/aptos2019-blindness-detection/train_images/5c7ab966a3ee.png" # notice upper part
path="F:\kaggleDataSet\diabeticRetinopathy\resized train 19\cd54d022e37d.jpg" # lower-right, this still looks not so severe, can be class3
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
height, width = image.shape
print(height, width)
SCALE=2
figsize = (width / float(dpi))/SCALE, (height / float(dpi))/SCALE
fig = plt.figure(figsize=figsize)
plt.imshow(image, cmap='gray')
%%time
fig = plt.figure(figsize=(25, 16))
for class_id in sorted(train_y.unique()):
for i, (idx, row) in enumerate(df_train.loc[df_train['diagnosis'] == class_id].sample(5, random_state=SEED).iterrows()):
ax = fig.add_subplot(5, 5, class_id * 5 + i + 1, xticks=[], yticks=[])
path="F:\kaggleDataSet\diabeticRetinopathy\resized train 19\"+str(row['id_code'])+".jpg"
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
# image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.resize(image, (IMG_SIZE, IMG_SIZE))
image=cv2.addWeighted ( image,4, cv2.GaussianBlur( image , (0,0) , IMG_SIZE/10) ,-4 ,128) # the trick is to add this line
plt.imshow(image, cmap='gray')
ax.set_title('Label: %d-%d-%s' % (class_id, idx, row['id_code']) )

def crop_image1(img,tol=7):
# img is image data
# tol is tolerance
mask = img>tol
return img[np.ix_(mask.any(1),mask.any(0))]
def crop_image_from_gray(img,tol=7):
if img.ndim ==2:
mask = img>tol
return img[np.ix_(mask.any(1),mask.any(0))]
elif img.ndim==3:
gray_img = cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)
mask = gray_img>tol
check_shape = img[:,:,0][np.ix_(mask.any(1),mask.any(0))].shape[0]
if (check_shape == 0): # image is too dark so that we crop out everything,
return img # return original image
else:
img1=img[:,:,0][np.ix_(mask.any(1),mask.any(0))]
img2=img[:,:,1][np.ix_(mask.any(1),mask.any(0))]
img3=img[:,:,2][np.ix_(mask.any(1),mask.any(0))]
# print(img1.shape,img2.shape,img3.shape)
img = np.stack([img1,img2,img3],axis=-1)
# print(img.shape)
return img
def load_ben_color(path, sigmaX=10):
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = crop_image_from_gray(image)
image = cv2.resize(image, (IMG_SIZE, IMG_SIZE))
image=cv2.addWeighted ( image,4, cv2.GaussianBlur( image , (0,0) , sigmaX) ,-4 ,128)
return image
%%time
NUM_SAMP=7
fig = plt.figure(figsize=(25, 16))
for class_id in sorted(train_y.unique()):
for i, (idx, row) in enumerate(df_train.loc[df_train['diagnosis'] == class_id].sample(NUM_SAMP, random_state=SEED).iterrows()):
ax = fig.add_subplot(5, NUM_SAMP, class_id * NUM_SAMP + i + 1, xticks=[], yticks=[])
path="F:\kaggleDataSet\diabeticRetinopathy\resized train 19\"+str(row['id_code'])+".jpg"
image = load_ben_color(path,sigmaX=30)
plt.imshow(image)
ax.set_title('%d-%d-%s' % (class_id, idx, row['id_code']) )

def circle_crop(img, sigmaX=10):
"""
Create circular crop around image centre
"""
img = cv2.imread(img)
img = crop_image_from_gray(img)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
height, width, depth = img.shape
x = int(width/2)
y = int(height/2)
r = np.amin((x,y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x,y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image_from_gray(img)
img=cv2.addWeighted ( img,4, cv2.GaussianBlur( img , (0,0) , sigmaX) ,-4 ,128)
return img
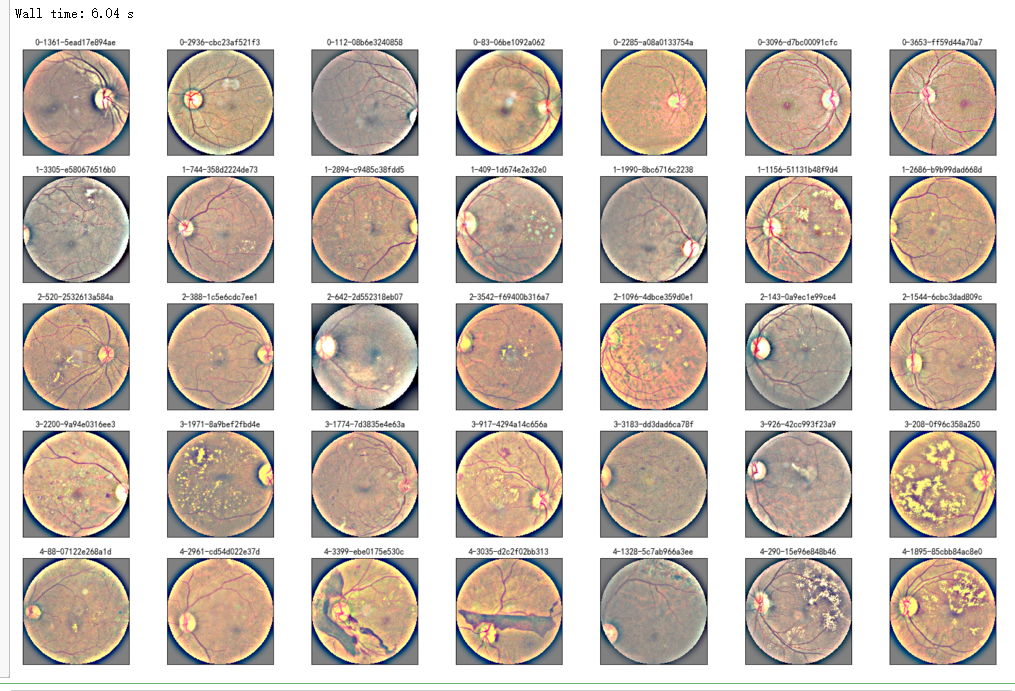
%%time
## try circle crop
NUM_SAMP=7
fig = plt.figure(figsize=(25, 16))
for class_id in sorted(train_y.unique()):
for i, (idx, row) in enumerate(df_train.loc[df_train['diagnosis'] == class_id].sample(NUM_SAMP, random_state=SEED).iterrows()):
ax = fig.add_subplot(5, NUM_SAMP, class_id * NUM_SAMP + i + 1, xticks=[], yticks=[])
path="F:\kaggleDataSet\diabeticRetinopathy\resized train 19\"+str(row['id_code'])+".jpg"
image = circle_crop(path,sigmaX=30)
plt.imshow(image)
ax.set_title('%d-%d-%s' % (class_id, idx, row['id_code']) )
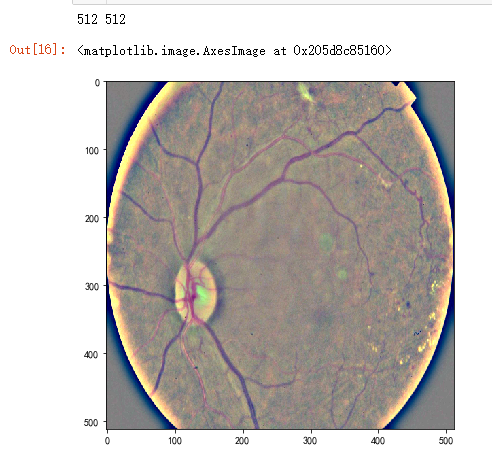
dpi = 80 #inch
path="F:\kaggleDataSet\diabeticRetinopathy\resized train 19\cd54d022e37d.jpg"
image = load_ben_color(path,sigmaX=10)
height, width = IMG_SIZE, IMG_SIZE
print(height, width)
SCALE=1
figsize = (width / float(dpi))/SCALE, (height / float(dpi))/SCALE
fig = plt.figure(figsize=figsize)
plt.imshow(image, cmap='gray')
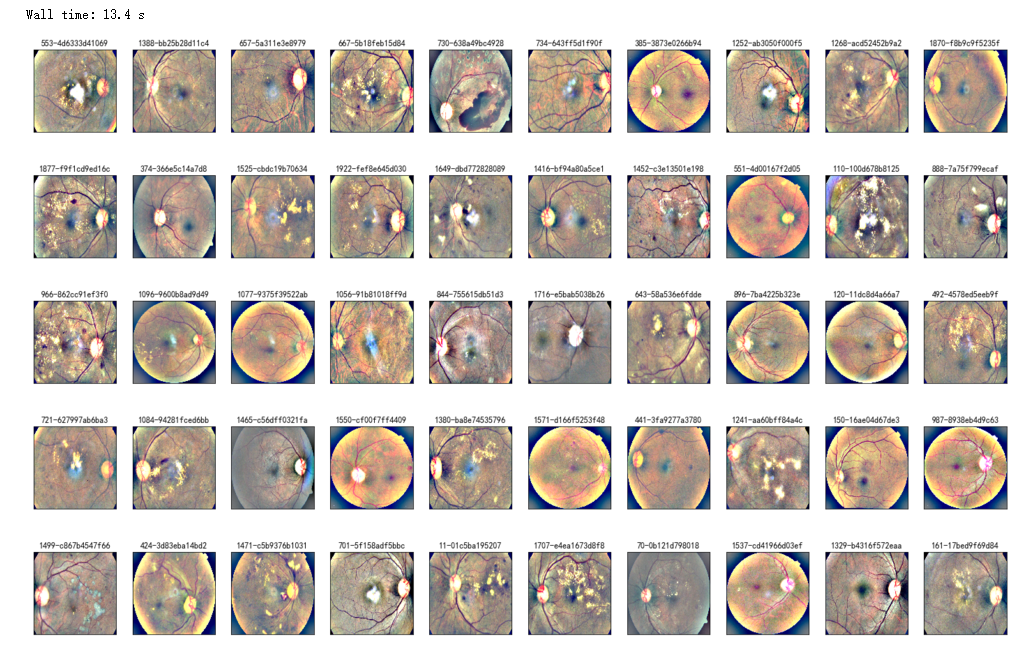
%%time
NUM_SAMP=10
fig = plt.figure(figsize=(25, 16))
for jj in range(5):
for i, (idx, row) in enumerate(df_test.sample(NUM_SAMP,random_state=SEED+jj).iterrows()):
ax = fig.add_subplot(5, NUM_SAMP, jj * NUM_SAMP + i + 1, xticks=[], yticks=[])
path="F:\kaggleDataSet\diabeticRetinopathy\resized test 19\"+str(row['id_code'])+".jpg"
image = load_ben_color(path,sigmaX=30)
plt.imshow(image)
ax.set_title('%d-%s' % (idx, row['id_code']) )

%%time
NUM_SAMP=10
fig = plt.figure(figsize=(25, 16))
for jj in range(5):
for i, (idx, row) in enumerate(df_test.sample(NUM_SAMP,random_state=SEED+jj).iterrows()):
ax = fig.add_subplot(5, NUM_SAMP, jj * NUM_SAMP + i + 1, xticks=[], yticks=[])
path="F:\kaggleDataSet\diabeticRetinopathy\resized test 19\"+str(row['id_code'])+".jpg"
image = load_ben_color(path,sigmaX=50)
plt.imshow(image, cmap='gray')
ax.set_title('%d-%s' % (idx, row['id_code']) )
df_old = pd.read_csv('F:\kaggleDataSet\diabeticRetinopathy\trainLabels.csv')
df_old.head()
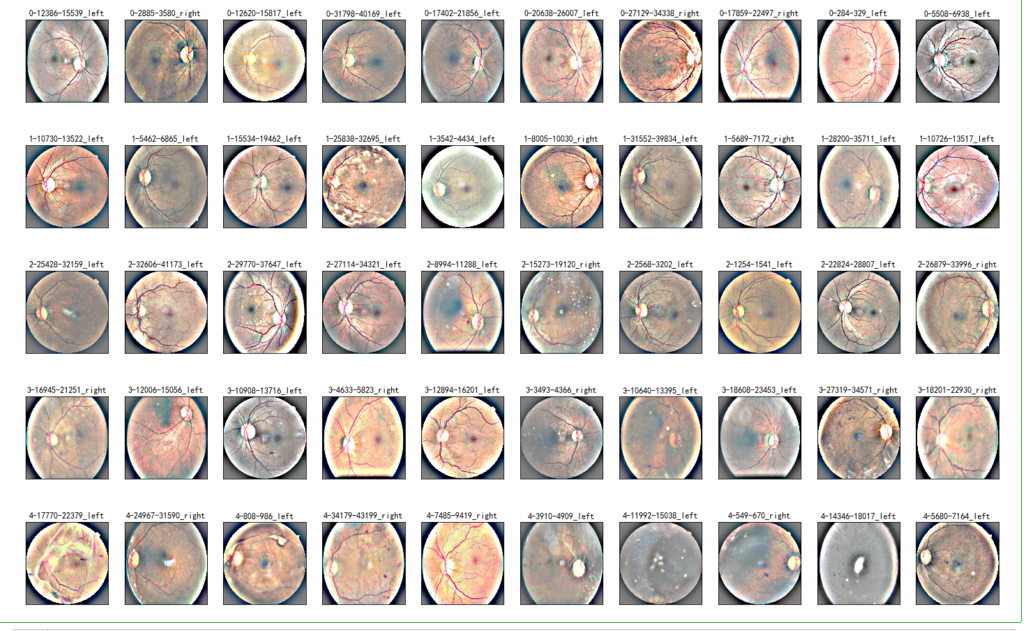
NUM_SAMP=10
fig = plt.figure(figsize=(25, 16))
for class_id in sorted(train_y.unique()):
for i, (idx, row) in enumerate(df_old.loc[df_old['level'] == class_id].sample(NUM_SAMP, random_state=SEED).iterrows()):
ax = fig.add_subplot(5, NUM_SAMP, class_id * NUM_SAMP + i + 1, xticks=[], yticks=[])
path="F:\kaggleDataSet\diabeticRetinopathy\resized_train\"+row['image']+".jpeg"
image = load_ben_color(path,sigmaX=30)
plt.imshow(image)
ax.set_title('%d-%d-%s' % (class_id, idx, row['image']) )
NUM_SAMP=10
fig = plt.figure(figsize=(25, 16))
for class_id in sorted(train_y.unique()):
for i, (idx, row) in enumerate(df_old.loc[df_old['level'] == class_id].sample(NUM_SAMP, random_state=SEED).iterrows()):
ax = fig.add_subplot(5, NUM_SAMP, class_id * NUM_SAMP + i + 1, xticks=[], yticks=[])
path="F:\kaggleDataSet\diabeticRetinopathy\resized_train\"+row['image']+".jpeg"
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.resize(image, (IMG_SIZE, IMG_SIZE))
plt.imshow(image, cmap='gray')
ax.set_title('%d-%d-%s' % (class_id, idx, row['image']) )

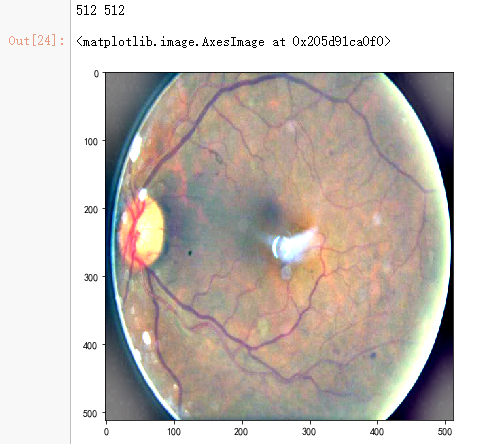
dpi = 80 #inch
path=f"F:\kaggleDataSet\diabeticRetinopathy\resized_train\31590_right.jpeg" # too many vessels?
image = load_ben_color(path,sigmaX=30)
# image = cv2.imread(path)
# image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
# image = crop_image1(image)
# image = cv2.resize(image, (IMG_SIZE, IMG_SIZE))
# image=cv2.addWeighted ( image,4, cv2.GaussianBlur( image , (0,0) , IMG_SIZE/10) ,-4 ,128)
height, width = IMG_SIZE, IMG_SIZE
print(height, width)
SCALE=1
figsize = (width / float(dpi))/SCALE, (height / float(dpi))/SCALE
fig = plt.figure(figsize=figsize)
plt.imshow(image, cmap='gray')
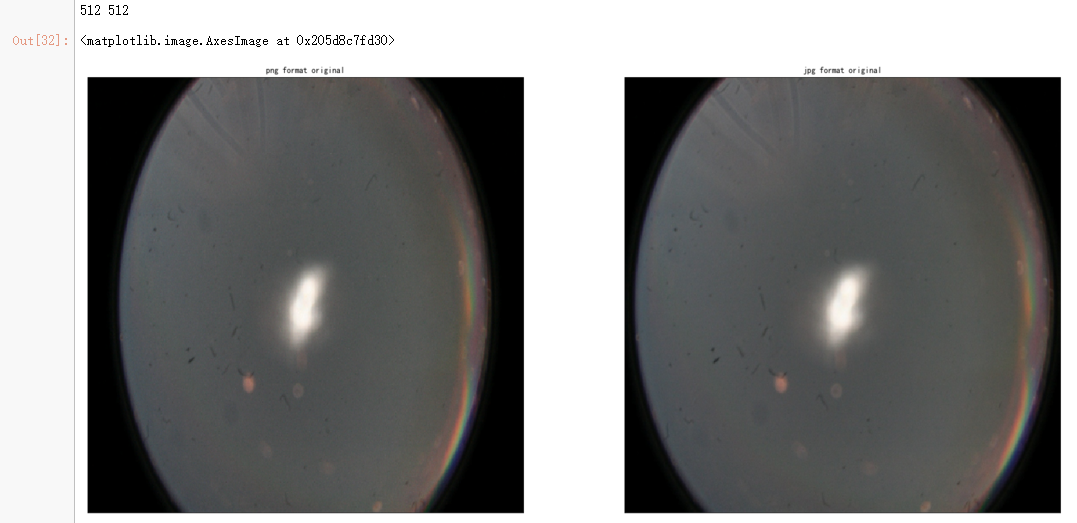
dpi = 80 #inch
path_jpg=f"F:\kaggleDataSet\diabeticRetinopathy\resized_train_cropped\18017_left.jpeg" # too many vessels?
path_png=f"F:\kaggleDataSet\diabeticRetinopathy\rescaled_train_896\18017_left.png" # details are lost
image = cv2.imread(path_png)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.resize(image, (IMG_SIZE, IMG_SIZE))
image2 = cv2.imread(path_jpg)
image2 = cv2.cvtColor(image2, cv2.COLOR_BGR2RGB)
image2 = cv2.resize(image2, (IMG_SIZE, IMG_SIZE))
height, width = IMG_SIZE, IMG_SIZE
print(height, width)
SCALE=1/4
figsize = (width / float(dpi))/SCALE, (height / float(dpi))/SCALE
fig = plt.figure(figsize=figsize)
ax = fig.add_subplot(2, 2, 1, xticks=[], yticks=[])
ax.set_title('png format original' )
plt.imshow(image, cmap='gray')
ax = fig.add_subplot(2, 2, 2, xticks=[], yticks=[])
ax.set_title('jpg format original' )
plt.imshow(image2, cmap='gray')
image = load_ben_color(path_png,sigmaX=30)
image2 = load_ben_color(path_jpg,sigmaX=30)
ax = fig.add_subplot(2, 2, 3, xticks=[], yticks=[])
ax.set_title('png format transformed' )
plt.imshow(image, cmap='gray')
ax = fig.add_subplot(2, 2, 4, xticks=[], yticks=[])
ax.set_title('jpg format transformed' )
plt.imshow(image2, cmap='gray')

import json
import math
import os
import cv2
from PIL import Image
import numpy as np
from keras import layers
from keras.applications import DenseNet121
from keras.callbacks import Callback, ModelCheckpoint
from keras.preprocessing.image import ImageDataGenerator
from keras.models import Sequential
from keras.optimizers import Adam
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.metrics import cohen_kappa_score, accuracy_score
import scipy
from tqdm import tqdm
%matplotlib inline
train_df = pd.read_csv('F:\kaggleDataSet\diabeticRetinopathy\trainLabels19.csv')
test_df = pd.read_csv('F:\kaggleDataSet\diabeticRetinopathy\testImages19.csv')
print(train_df.shape)
print(test_df.shape)
test_df.head()
def get_pad_width(im, new_shape, is_rgb=True):
pad_diff = new_shape - im.shape[0], new_shape - im.shape[1]
t, b = math.floor(pad_diff[0]/2), math.ceil(pad_diff[0]/2)
l, r = math.floor(pad_diff[1]/2), math.ceil(pad_diff[1]/2)
if is_rgb:
pad_width = ((t,b), (l,r), (0, 0))
else:
pad_width = ((t,b), (l,r))
return pad_width
def preprocess_image(image_path, desired_size=224):
im = Image.open(image_path)
im = im.resize((desired_size, )*2, resample=Image.LANCZOS)
return im
N = test_df.shape[0]
x_test = np.empty((N, 224, 224, 3), dtype=np.uint8)
for i, image_id in enumerate(tqdm(test_df['id_code'])):
x_test[i, :, :, :] = preprocess_image("F:\kaggleDataSet\diabeticRetinopathy\resized test 19\"+str(image_id)+".jpg")

# model.summary()
def load_image_ben_orig(path,resize=True,crop=False,norm255=True,keras=False):
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image=cv2.addWeighted( image,4, cv2.GaussianBlur( image , (0,0) , 10) ,-4 ,128)
if norm255:
return image/255
elif keras:
#see https://github.com/keras-team/keras-applications/blob/master/keras_applications/imagenet_utils.py for mode
#see https://github.com/keras-team/keras-applications/blob/master/keras_applications/xception.py for inception,xception mode
#the use of tf based preprocessing (- and / by 127 respectively) will results in [-1,1] so it will not visualize correctly (directly)
image = np.expand_dims(image, axis=0)
return preprocess_input(image)[0]
else:
return image.astype(np.int16)
return image
def transform_image_ben(img,resize=True,crop=False,norm255=True,keras=False):
image=cv2.addWeighted( img,4, cv2.GaussianBlur( img , (0,0) , 10) ,-4 ,128)
if norm255:
return image/255
elif keras:
image = np.expand_dims(image, axis=0)
return preprocess_input(image)[0]
else:
return image.astype(np.int16)
return image
def display_samples(df, columns=5, rows=2, Ben=True):
fig=plt.figure(figsize=(5*columns, 4*rows))
for i in range(columns*rows):
image_path = df.loc[i,'id_code']
path = f"F:\kaggleDataSet\diabeticRetinopathy\resized test 19\"+str(image_path)+".jpg"
if Ben:
img = load_image_ben_orig(path)
else:
img = cv2.imread(path)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
fig.add_subplot(rows, columns, i+1)
plt.imshow(img)
plt.tight_layout()
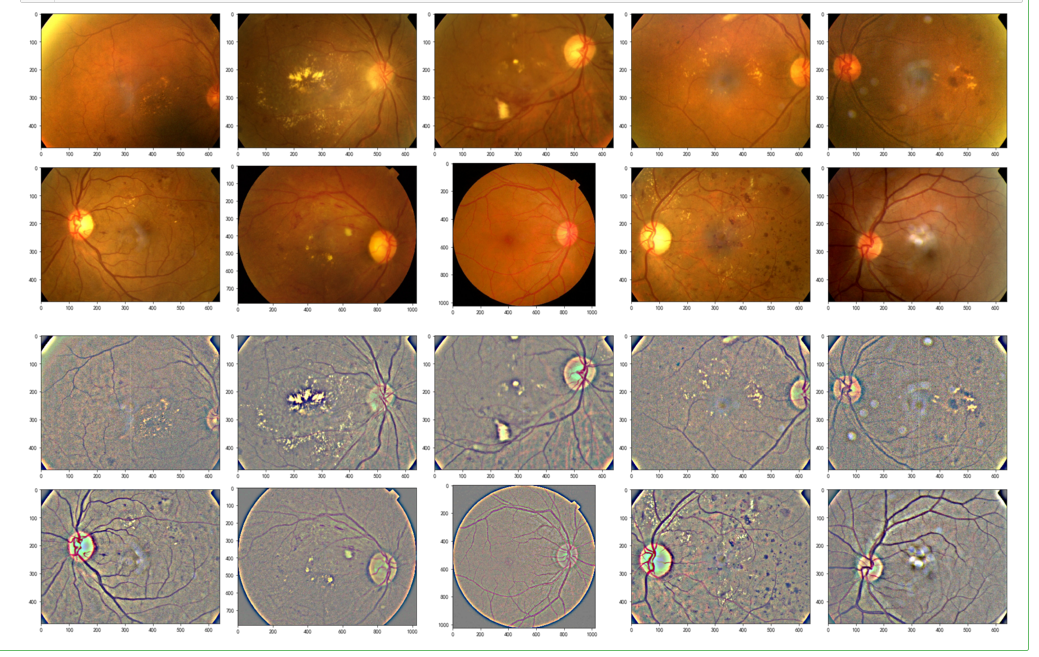
display_samples(test_df, Ben=False)
display_samples(test_df, Ben=True)
from keras import layers
from keras.models import Model
import keras.backend as K
K.clear_session()
densenet = DenseNet121(weights=None,include_top=False,input_shape=(None,None,3))
GAP_layer = layers.GlobalAveragePooling2D()
drop_layer = layers.Dropout(0.5)
dense_layer = layers.Dense(5, activation='sigmoid', name='final_output')
def build_model_sequential():
model = Sequential()
model.add(densenet)
model.add(GAP_layer)
model.add(drop_layer)
model.add(dense_layer)
return model
modelA = build_model_sequential()
modelA.load_weights('F:\kaggleDataSet\diabeticRetinopathy\dense_xhlulu_731.h5')
modelA.summary()
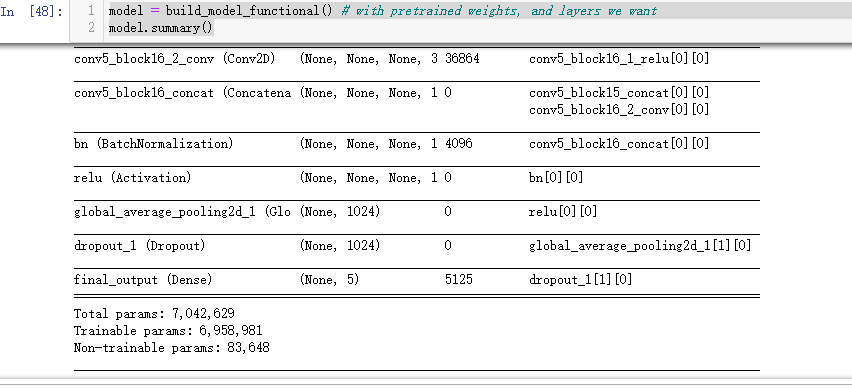
model = build_model_functional() # with pretrained weights, and layers we want
model.summary()
y_test = model.predict(x_test) > 0.5
y_test = y_test.astype(int).sum(axis=1) - 1
import seaborn as sns
import cv2
SIZE=224
def create_pred_hist(pred_level_y,title='NoTitle'):
results = pd.DataFrame({'diagnosis':pred_level_y})
f, ax = plt.subplots(figsize=(7, 4))
ax = sns.countplot(x="diagnosis", data=results, palette="GnBu_d")
sns.despine()
plt.title(title)
plt.show()
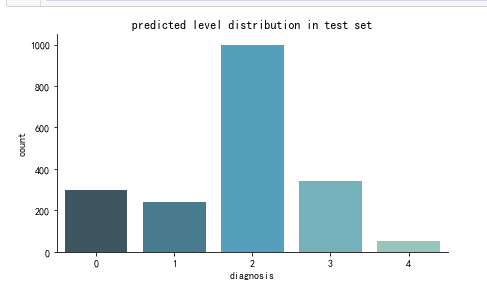
create_pred_hist(y_test,title='predicted level distribution in test set')
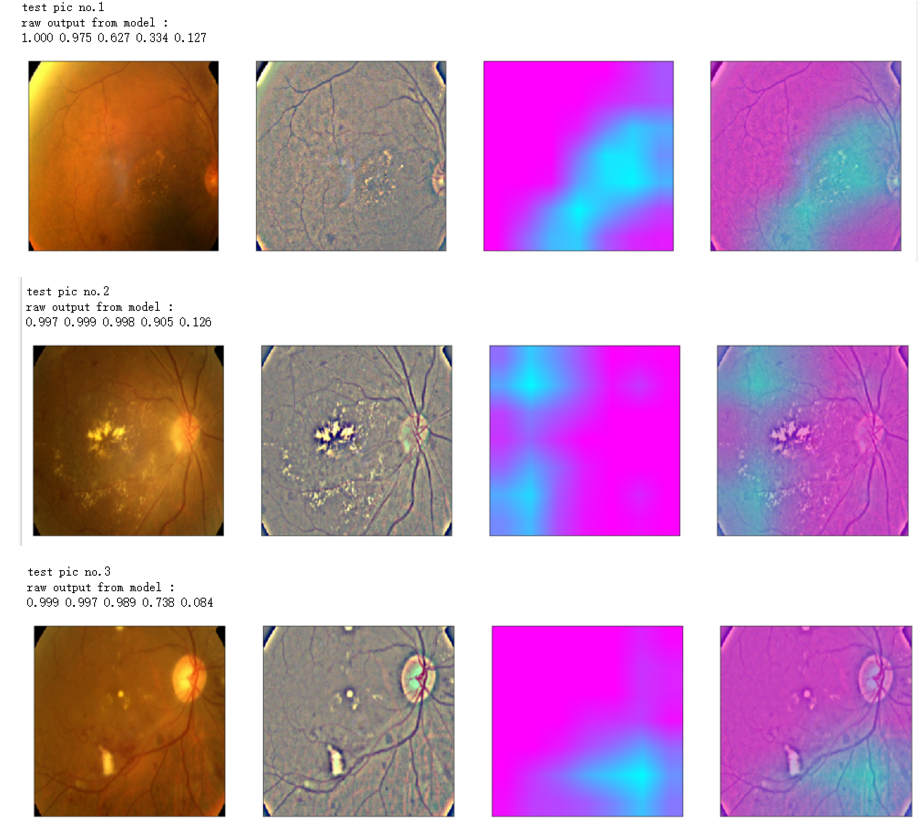
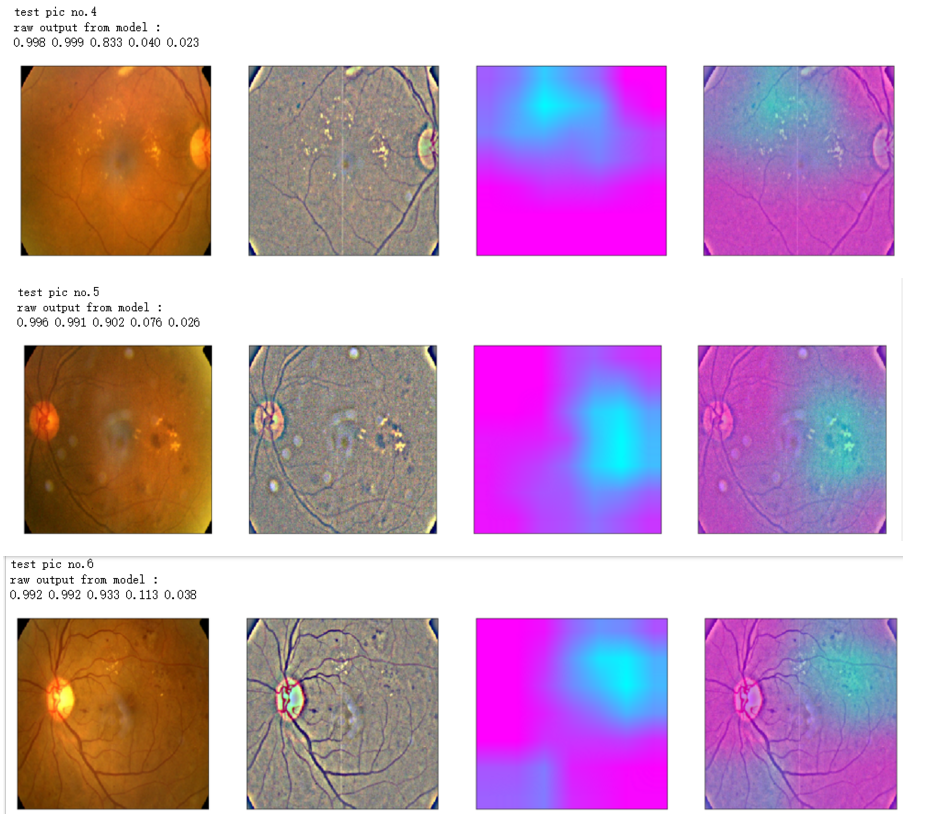
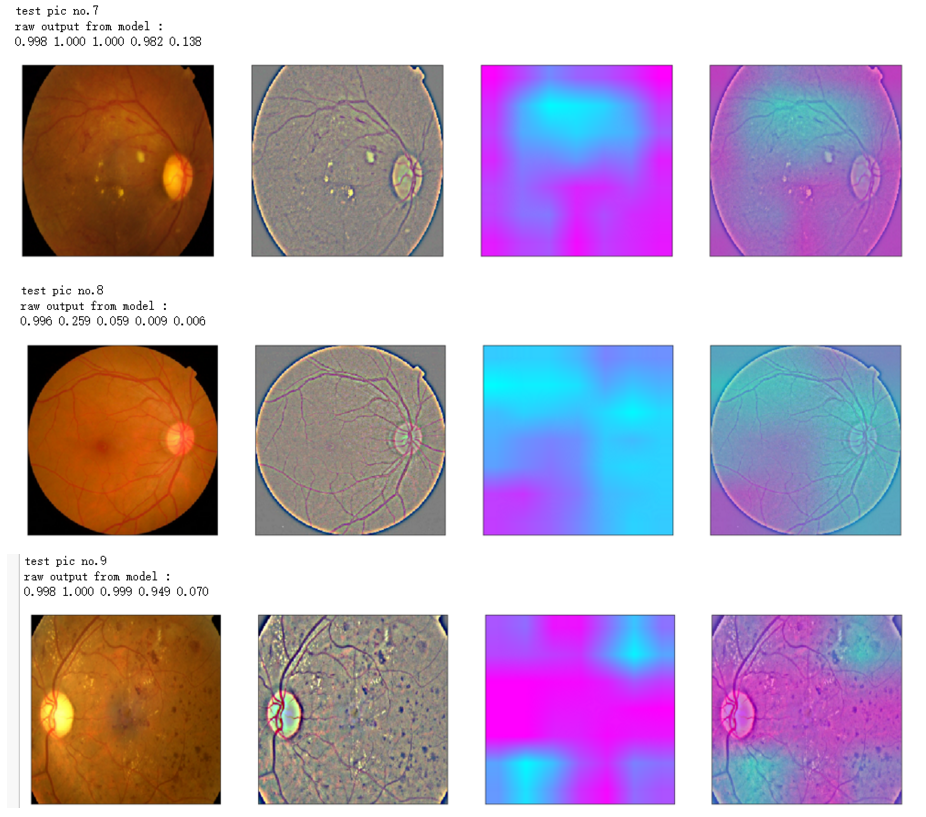
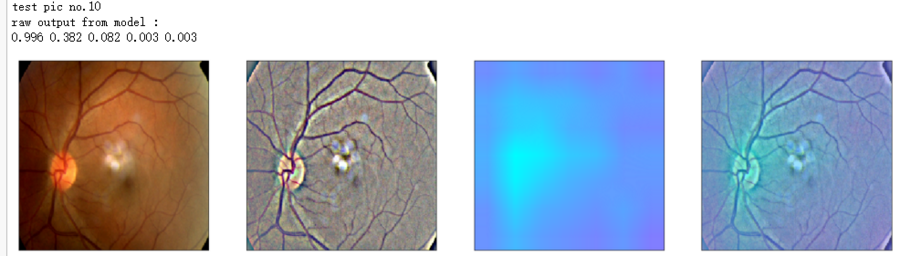
def gen_heatmap_img(img, model0, layer_name='last_conv_layer',viz_img=None,orig_img=None):
preds_raw = model0.predict(img[np.newaxis])
preds = preds_raw > 0.5 # use the same threshold as @xhlulu original kernel
class_idx = (preds.astype(int).sum(axis=1) - 1)[0]
class_output_tensor = model0.output[:, class_idx]
viz_layer = model0.get_layer(layer_name)
grads = K.gradients(class_output_tensor ,viz_layer.output)[0] # gradients of viz_layer wrt output_tensor of predicted class
pooled_grads=K.mean(grads,axis=(0,1,2))
iterate=K.function([model0.input],[pooled_grads, viz_layer.output[0]])
pooled_grad_value, viz_layer_out_value = iterate([img[np.newaxis]])
for i in range(pooled_grad_value.shape[0]):
viz_layer_out_value[:,:,i] *= pooled_grad_value[i]
heatmap = np.mean(viz_layer_out_value, axis=-1)
heatmap = np.maximum(heatmap,0)
heatmap /= np.max(heatmap)
viz_img=cv2.resize(viz_img,(img.shape[1],img.shape[0]))
heatmap=cv2.resize(heatmap,(viz_img.shape[1],viz_img.shape[0]))
heatmap_color = cv2.applyColorMap(np.uint8(heatmap*255), cv2.COLORMAP_SPRING)/255
heated_img = heatmap_color*0.5 + viz_img*0.5
print('raw output from model : ')
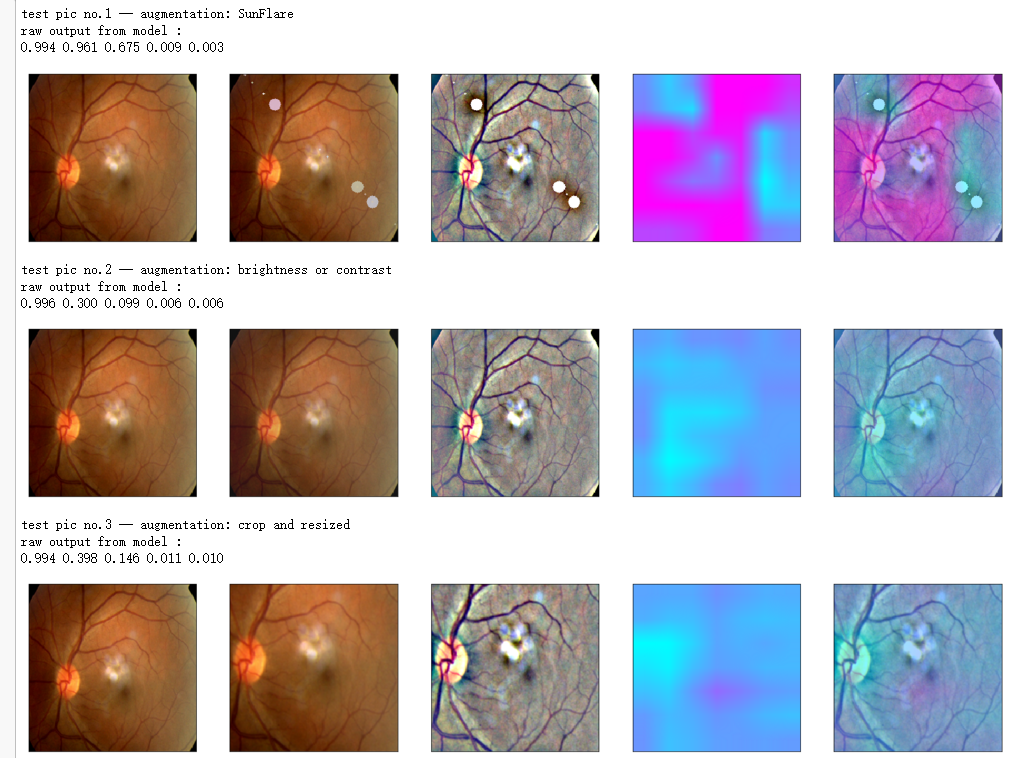
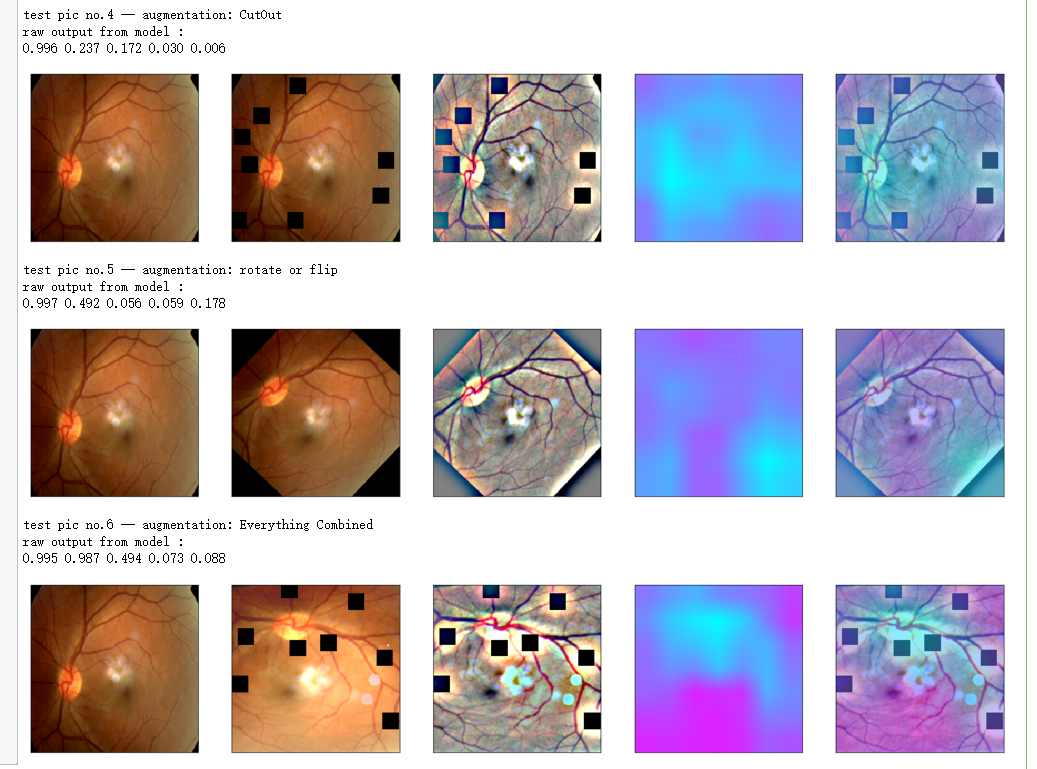
print_pred(preds_raw)
if orig_img is None:
show_Nimages([img,viz_img,heatmap_color,heated_img])
else:
show_Nimages([orig_img,img,viz_img,heatmap_color,heated_img])
plt.show()
return heated_img
def show_image(image,figsize=None,title=None):
if figsize is not None:
fig = plt.figure(figsize=figsize)
if image.ndim == 2:
plt.imshow(image,cmap='gray')
else:
plt.imshow(image)
if title is not None:
plt.title(title)
def show_Nimages(imgs,scale=1):
N=len(imgs)
fig = plt.figure(figsize=(25/scale, 16/scale))
for i, img in enumerate(imgs):
ax = fig.add_subplot(1, N, i + 1, xticks=[], yticks=[])
show_image(img)
def print_pred(array_of_classes):
xx = array_of_classes
s1,s2 = xx.shape
for i in range(s1):
for j in range(s2):
print('%.3f ' % xx[i,j],end='')
print('')
NUM_SAMP=10
SEED=77
layer_name = 'relu' #'conv5_block16_concat'
for i, (idx, row) in enumerate(test_df[:NUM_SAMP].iterrows()):
path=f"F:\kaggleDataSet\diabeticRetinopathy\resized test 19\"+str(row["id_code"])+".jpg"
ben_img = load_image_ben_orig(path)
input_img = np.empty((1,224, 224, 3), dtype=np.uint8)
input_img[0,:,:,:] = preprocess_image(path)
print('test pic no.%d' % (i+1))
_ = gen_heatmap_img(input_img[0],model, layer_name=layer_name,viz_img=ben_img)
from albumentations import *
import time
IMG_SIZE = (224,224)
'''Use case from https://www.kaggle.com/alexanderliao/image-augmentation-demo-with-albumentation/'''
def albaugment(aug0, img):
return aug0(image=img)['image']
idx=8
image1=x_test[idx]
'''1. Rotate or Flip'''
aug1 = OneOf([Rotate(p=0.99, limit=160, border_mode=0,value=0), Flip(p=0.5)],p=1)
'''2. Adjust Brightness or Contrast'''
aug2 = RandomBrightnessContrast(brightness_limit=0.45, contrast_limit=0.45,p=1)
h_min=np.round(IMG_SIZE[1]*0.72).astype(int)
h_max= np.round(IMG_SIZE[1]*0.9).astype(int)
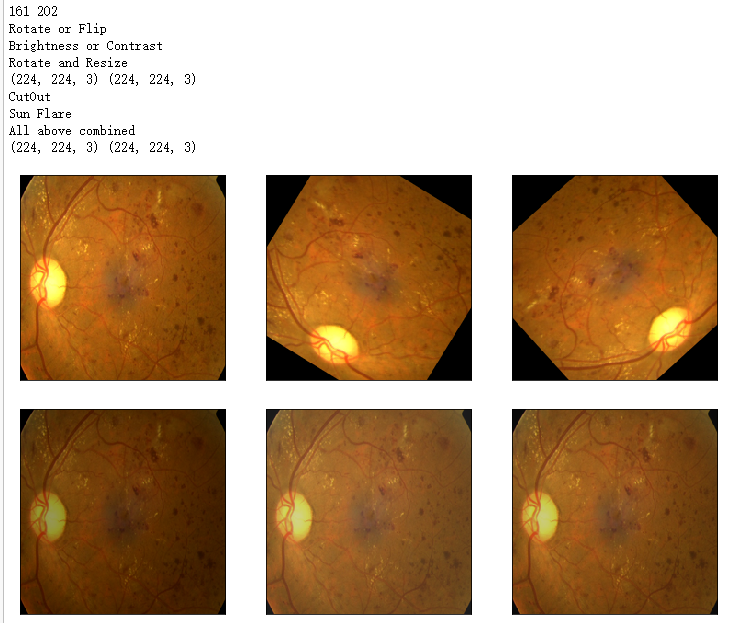
print(h_min,h_max)
'''3. Random Crop and then Resize'''
#w2h_ratio = aspect ratio of cropping
aug3 = RandomSizedCrop((h_min, h_max),IMG_SIZE[1],IMG_SIZE[0], w2h_ratio=IMG_SIZE[0]/IMG_SIZE[1],p=1)
'''4. CutOut Augmentation'''
max_hole_size = int(IMG_SIZE[1]/10)
aug4 = Cutout(p=1,max_h_size=max_hole_size,max_w_size=max_hole_size,num_holes=8 )#default num_holes=8
'''5. SunFlare Augmentation'''
aug5 = RandomSunFlare(src_radius=max_hole_size,num_flare_circles_lower=10,num_flare_circles_upper=20,p=1)
'''6. Ultimate Augmentation -- combine everything'''
final_aug = Compose([aug1,aug2,aug3,aug4,aug5],p=1)
img1 = albaugment(aug1,image1)
img2 = albaugment(aug1,image1)
print('Rotate or Flip')
show_Nimages([image1,img1,img2],scale=2)
# time.sleep(1)
img1 = albaugment(aug2,image1)
img2 = albaugment(aug2,image1)
img3 = albaugment(aug2,image1)
print('Brightness or Contrast')
show_Nimages([img3,img1,img2],scale=2)
# time.sleep(1)
img1 = albaugment(aug3,image1)
img2 = albaugment(aug3,image1)
img3 = albaugment(aug3,image1)
print('Rotate and Resize')
show_Nimages([img3,img1,img2],scale=2)
print(img1.shape,img2.shape)
# time.sleep(1)
img1 = albaugment(aug4,image1)
img2 = albaugment(aug4,image1)
img3 = albaugment(aug4,image1)
print('CutOut')
show_Nimages([img3,img1,img2],scale=2)
# time.sleep(1)
img1 = albaugment(aug5,image1)
img2 = albaugment(aug5,image1)
img3 = albaugment(aug5,image1)
print('Sun Flare')
show_Nimages([img3,img1,img2],scale=2)
# time.sleep(1)
img1 = albaugment(final_aug,image1)
img2 = albaugment(final_aug,image1)
img3 = albaugment(final_aug,image1)
print('All above combined')
show_Nimages([img3,img1,img2],scale=2)
print(img1.shape,img2.shape)
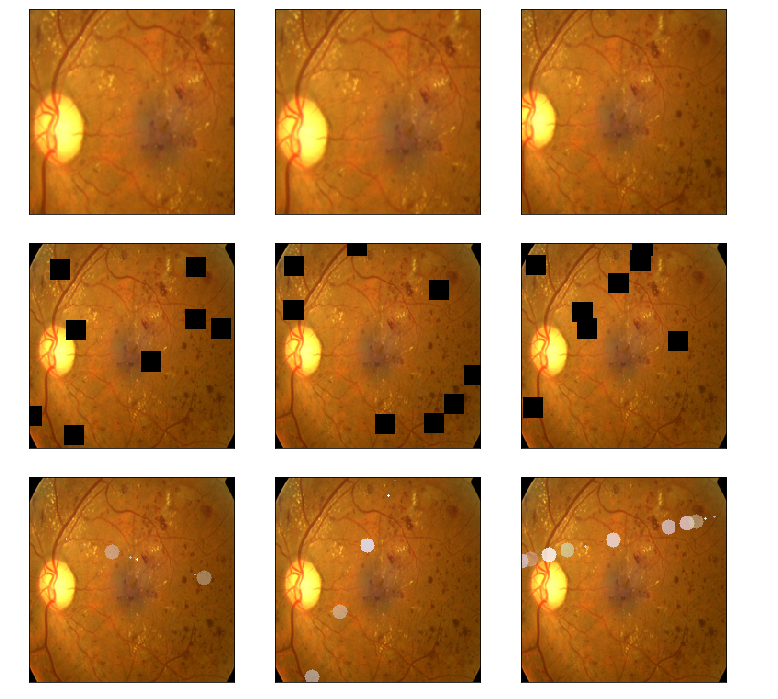
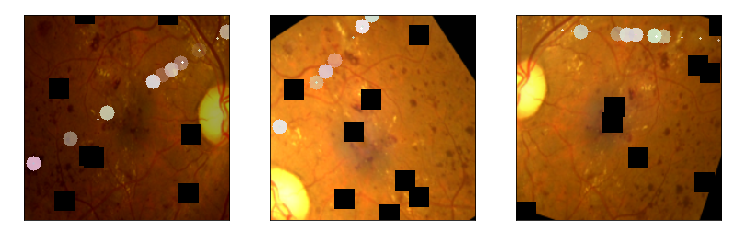
aug_list = [aug5, aug2, aug3, aug4, aug1, final_aug]
aug_name = ['SunFlare', 'brightness or contrast', 'crop and resized', 'CutOut', 'rotate or flip', 'Everything Combined']
idx=8
layer_name = 'relu'
for i in range(len(aug_list)):
path=f"F:\kaggleDataSet\diabeticRetinopathy\resized test 19\"+str(row["id_code"])+".jpg"
input_img = np.empty((1,224, 224, 3), dtype=np.uint8)
input_img[0,:,:,:] = preprocess_image(path)
aug_img = albaugment(aug_list[i],input_img[0,:,:,:])
ben_img = transform_image_ben(aug_img)
print('test pic no.%d -- augmentation: %s' % (i+1, aug_name[i]))
_ = gen_heatmap_img(aug_img,model, layer_name=layer_name,viz_img=ben_img,orig_img=input_img[0])