logistic regression是分类算法中非常重要的算法,也是非常基础的算法。logistic regression从整体上考虑样本预测的精度,用判别学习模型的条件似然进行参数估计,假设样本遵循iid,参数估计时保证每个样本的预测值接近真实值的概率最大化。这样的结果,只能是牺牲一部分的精度来换取另一部分的精度。而svm从局部出发,假设有一个分类平面,找出所有距离分类平面的最近的点(support vector,数量很少),让这些点到平面的距离最大化,那么这个分类平面就是最佳分类平面。从这个角度来看待两个算法,可以得出logistic regression的精度肯定要低于后者。今天主要写logistic regression的Python代码。logistic regression的推导过程比较简单:
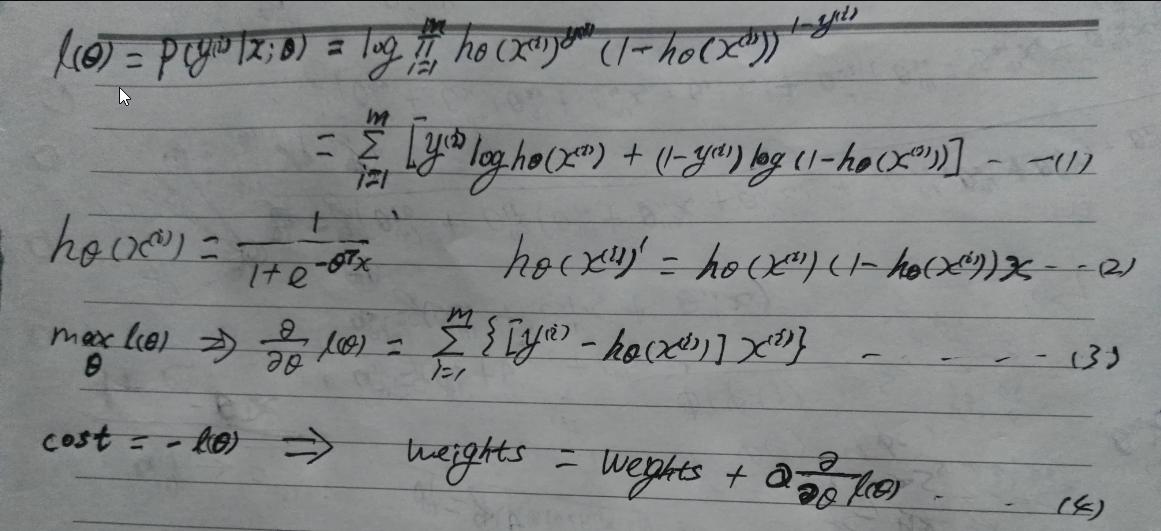
第一个公式是条件似然函数估计,意思是指定未知常量theta(;表示频率学派),对于每个输入feature vector x(i),产生y(i)的概率都最大,取对数是为了求导方便。第二个公式是sigmoid函数的导数,在这里推导出具体的导函数(推导过程非常简单,复合函数求导法则),第三个公式是求出的梯度(实际为偏导数组成的向量,梯度与方向导数同方向时取得最大值,相反时取得最小值)。公式3的线性意义为:对于容量为m的样本(矩阵mxn),权重为nx1的列向量,每一行的样本数据与权重向量相乘求得预测值,m行样本的预测值组成mx1的列向量(其数值为exp(-inX)),其实就是mxn的样本矩阵左乘权重向量nx1,因为的矩阵的本质是线性变换,相当于每一行样本数据投影到权重向量。得到预测值的列向量后,与真实值向量(需要转置)做差值得到新的列向量。最后样本矩阵倒置,然后左乘这个向量得到nx1的权重向量(梯度)。这个过程是计算批量梯度。当似然函数取得最大时,就是损失函数最小,所以二者是相反的关系,最后结果也就是梯度上升法。
对于逻辑回归的损失函数,也可以从另外一个角度来理解。上面的公式我们看到损失函数和最大似然函数是相反的关系,一般情况下,logistic regression的loss function 可以采用交叉熵的形式,然后取mean。
SGD(stochastic gradient descent)在计算的过程中,每次迭代不用计算所用样本,而是随机选取一个样本进行梯度上升(下降),在工程实践中可以满足精度要求,而且时间复杂度比批量要低。对于SGD而言,学习速率alpha的选取,随着循环次数增加,刚开始的时候,梯度下降应该比较快,到后期的时候,可能会出现在某一个值徘徊的情况,而且下降速率会越来越慢。所以选取一个比较合理的study rate,应该是先选取到的样本study rate相对较高,后面的相对较小,这样比较合理。
在训练出模型后,预测时应该使用"五折交叉验证理论"寻找出最优模型出来(调节参数的过程),这个过程一般用RMSE(均方根误差)来衡量,并且可以定义一个基准均方根误差。在用最优模型预测时取得的RMSE与基准RMSE对比,分析数据结果。
本文主要探讨logistic regression的SGD算法,并且不考虑正则化。事实上,在高维度的情况下,泛化能力或者正则化技术非常关键。在90年代有学者提出lasso后,至今有很多方法实践了lasso,比如LARS,CD……。下一篇博客将探讨lasso技术,并且动手实践CD算法。接下来,上传最近写的SGD Python代码,首先是引入模块:logisticRegression.py,这里面定义了两个class:LogisticRegressionWithSGD,LRModel,还有全局函数RMSE,loadDataSet和sigmoid函数。后面是测试代码,主要是参数调优。
logisticRegression.py:

1 ''' 2 Created on 2017/03/15 3 Logistic Regression without lasso 4 @author: XueQiang Tong 5 ''' 6 ''' 7 this algorithm include SGD,batch gradient descent except lasso regularization,So the generalization 8 ability is relatively weak, follow-up and then write a CD algorithm for lasso. 9 ''' 10 from numpy import * 11 import matplotlib.pyplot as plt 12 import numpy as np; 13 14 # load dataset into ndarray(numpy) 15 def loadDataSet(filepath, seperator=" "): 16 with open(filepath) as fr: 17 lines = fr.readlines(); 18 num_samples = len(lines); 19 dimension = len(lines[0].strip().split(seperator)); 20 dataMat = np.zeros((num_samples, dimension)); 21 labelMat = []; 22 23 index = 0; 24 for line in lines: 25 sample = line.strip().split(); 26 feature = list(map(np.float32, sample[:-1])); 27 dataMat[index, 1:] = feature; 28 dataMat[index, 0] = 1.0; 29 labelMat.append(float(sample[-1])); 30 index += 1; 31 32 return dataMat, array(labelMat); 33 34 #sigmoid function 35 def sigmoid(inX): 36 return 1.0/(1+exp(-inX)) 37 38 # compute rmse 39 def RMSE(true,predict): 40 true_predict = zip(true,predict); 41 sub = []; 42 for r,p in true_predict: 43 sub.append(math.pow((r-p),2)); 44 return math.sqrt(mean(sub)); 45 46 def maxstep(dataMat): 47 return 2 / abs(np.linalg.det(np.dot(dataMat.transpose(),dataMat))); 48 49 class LRModel: 50 def __init__(self,weights = np.empty(10),alpha = 0.01,iter = 150): 51 self.weights = weights; 52 self.alpha = alpha; 53 self.iter = iter; 54 55 #predict 56 def predict(self,inX): 57 prob = sigmoid(dot(inX,self.weights)) 58 if prob > 0.5: return 1.0 59 else: return 0.0 60 61 def getWeights(self): 62 return self.weights; 63 64 def __getattr__(self, item): 65 if item == 'weights': 66 return self.weights; 67 68 class LogisticRegressionWithSGD: 69 # batch gradient descent 70 @classmethod 71 def batchGradDescent(cls,data,maxCycles = 500,alpha = 0.001): 72 dataMatrix = mat(data[0]) # convert to NumPy matrix 73 labelMat = mat(data[1]).transpose() # convert to NumPy matrix 74 m,n = shape(dataMatrix) 75 weights = ones((n,1)) 76 for k in range(maxCycles): 77 h = sigmoid(dataMatrix*weights) # compute predict_value 78 error = (labelMat - h) # compute deviation 79 weights = weights + alpha * dataMatrix.transpose()* error # update weight 80 81 model = LRModel(weights = weights,alpha = alpha,iter = maxCycles); 82 return model; 83 84 ''' 85 # draw samples of the scatter plot to view the distribution of sample points 86 @classmethod 87 def viewScatterPlot(cls,weights,dataMat,labelMat): 88 dataArr = array(dataMat) 89 n = shape(dataArr)[0] 90 xcord1 = []; ycord1 = [] 91 xcord2 = []; ycord2 = [] 92 for i in range(n): 93 if int(labelMat[i])== 1: 94 xcord1.append(dataArr[i,1]); ycord1.append(dataArr[i,2]) 95 else: 96 xcord2.append(dataArr[i,1]); ycord2.append(dataArr[i,2]) 97 fig = plt.figure() 98 ax = fig.add_subplot(111) 99 ax.scatter(xcord1, ycord1, s=30, c='red', marker='s',label='1') 100 ax.scatter(xcord2, ycord2, s=30, c='green',label='0') 101 plt.legend(); 102 x = arange(-3.0, 3.0, 0.1) 103 y = (-weights[0]-weights[1]*x)/weights[2] 104 ax.plot(mat(x), mat(y)) 105 plt.xlabel('X1'); plt.ylabel('X2'); 106 plt.show() 107 ''' 108 @classmethod 109 def train_nonrandom(cls,data,alpha = 0.01): 110 dataMatrix = data[0]; 111 classLabels = data[1]; 112 m,n = shape(dataMatrix); 113 weights = ones(n) #initialize to all ones 114 for i in range(m): 115 h = sigmoid(sum(dataMatrix[i]*weights)); 116 error = classLabels[i] - h; 117 weights = weights + alpha * error * dataMatrix[i]; 118 119 model = LRModel(weights = weights,alpha = alpha); 120 return model; 121 122 @classmethod 123 def train_random(cls,data, numIter=150): 124 dataMatrix = data[0]; 125 classLabels = data[1]; 126 m,n = shape(dataMatrix); 127 weights = ones(n); #initialize to all ones 128 indices = np.arange(len(dataMatrix)) 129 for iter in range(numIter): 130 np.random.shuffle(indices) 131 for index in indices: 132 alpha = 4 / (1.0 + iter + index) + 0.0001 #apha decreases with iteration, does not 133 h = sigmoid(sum(dataMatrix[index]*weights)); 134 error = classLabels[index] - h; 135 weights = weights + alpha * error * array(dataMatrix[index]); 136 137 model = LRModel(weights = weights,iter = numIter); 138 return model; 139 140 def colicTest(self): 141 data = loadDataSet('G:\testSet.txt'); 142 trainWeights = self.stoGradDescent_random(data); 143 dataMat = data[0]; 144 labels = data[1]; 145 errnums = 0; 146 for index in range(dataMat.shape[0]): 147 preVal = self.predict(dataMat[index,:],trainWeights); 148 if(preVal != labels[index]): 149 errnums += 1; 150 print('error rate:%.2f' % (errnums/dataMat.shape[0])); 151 return errnums; 152 153 def multiTest(self): 154 numTests = 10; errorSum=0.0 155 for k in range(numTests): 156 errorSum += self.colicTest() 157 print("after %d iterations the average error rate is: %.2f" % (numTests, errorSum/numTests))
测试代码,把最优模型保存在npy文件里,以后使用的时候,直接取出来,不用再训练了。

1 from logisticRegression import *; 2 from numpy import *; 3 import math; 4 5 def main(): 6 dataMat, labels = loadDataSet('G:\testSet.txt'); 7 num_samples = dataMat.shape[0]; 8 9 num_trains = int(num_samples * 0.6); 10 num_validations = int(num_samples * 0.2); 11 num_tests = int(num_samples * 0.2); 12 13 data_trains = dataMat[:num_trains, :]; 14 data_validations = dataMat[num_trains:(num_trains + num_validations), :]; 15 data_tests = dataMat[(num_trains + num_validations):, :]; 16 17 label_trains = labels[:num_trains]; 18 label_validations = labels[num_trains:(num_trains + num_validations)]; 19 label_tests = labels[(num_trains + num_validations):]; 20 ''' 21 minrmse = (1 << 31) - 1; 22 bestModel = LRModel(); 23 iterList = [10, 20, 30, 80]; 24 25 for iter in iterList: 26 model = LogisticRegressionWithSGD.train_random((data_trains, label_trains), numIter=iter); 27 preVals = zeros(num_validations); 28 for i in range(num_validations): 29 preVals[i] = model.predict(data_validations[i, :]); 30 31 rmse = RMSE(label_validations, preVals); 32 if rmse < minrmse: 33 minrmse = rmse; 34 bestModel = model; 35 36 print(bestModel.iter, bestModel.weights, minrmse); 37 save('D:\Python\models\weights.npy',bestModel.weights);''' 38 39 #用最佳模型预测测试集的评分,并计算和实际评分之间的均方根误差 40 weights = load('D:\Python\models\weights.npy'); 41 42 LogisticRegressionWithSGD.viewScatterPlot(weights,dataMat,labels);#显示散点图 43 model = LRModel(); 44 model.weights = weights; 45 46 preVals = zeros(num_tests); 47 for i in range(num_tests): 48 preVals[i] = model.predict(data_tests[i,:]); 49 50 51 testRMSE = RMSE(label_tests,preVals); #预测产生的均方根误差 52 #用基准偏差衡量最佳模型在测试数据上的预测精度 53 tavMean = mean(hstack((label_trains,label_validations))); 54 baseRMSE = math.sqrt(mean((label_tests - tavMean) ** 2)) #基准均方根误差 55 56 improvement = abs(testRMSE - baseRMSE) / baseRMSE * 100; 57 58 print("The best model improves the base line by %% %1.2f" % (improvement)); 59 60 if __name__ == '__main__': 61 main();
运行结果:
样本点的散点图:
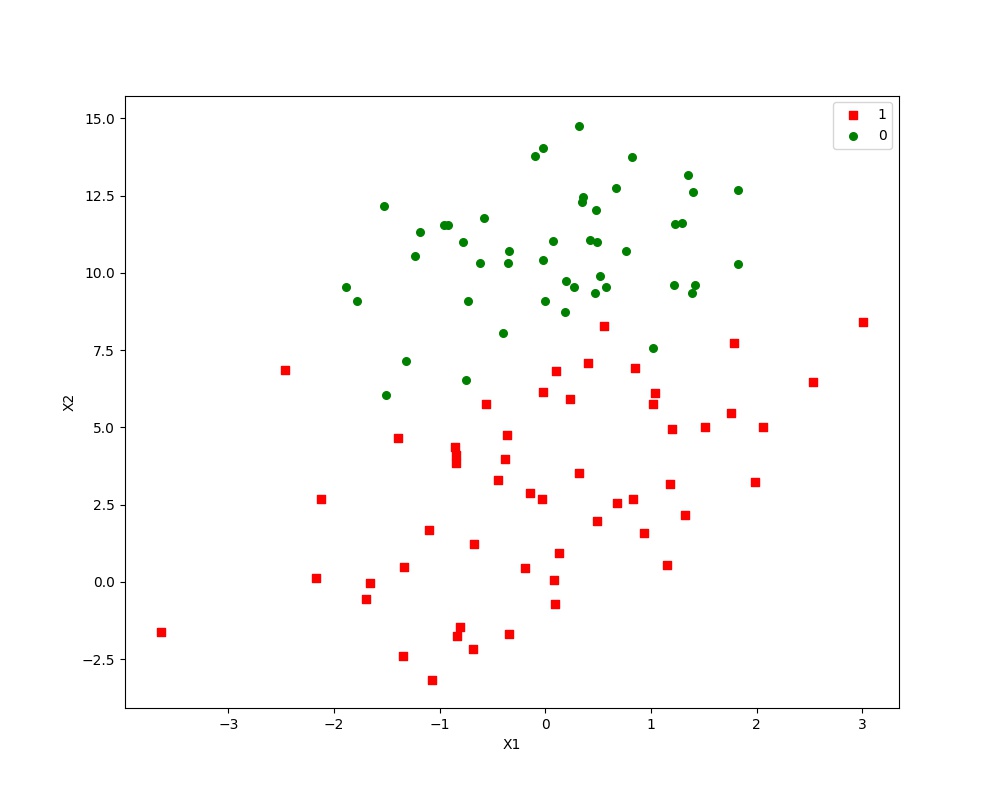
最佳迭代次数,权重以及RMSE: 10 [ 12.08509707 1.4723024 -1.86595103] 0.0
The best model improves the base line by % 55.07
另外,由于训练过程中是随机选取样本点,所以迭代次数相同的情况下,权重以及RMSE有可能不同,我们要的是RMSE最小的模型!
算法比较简单,但是用Python的nump库实施的时候,有很多注意的细节,只有经过自己仔细的理论推导然后再代码实施后,才能算基本掌握了一个算法。写代码及优化的过程是很费时的,后续还要改进算法,深化理论研究,并且坚持理论与编程结合,切不可眼高手低!
