1 推荐系统概述
1.1 从亚马逊网站认识推荐系统
首先,推荐系统的算法并不复杂。
从需求的定位上来看,在大多数的情况下,用户的需求很难用简单的关键字来表述,需求通常是来源于不完整的记忆,或者是道听途说等,很少有人能记住不熟悉的产品名称。在这种情况下,如果用传统的搜索引擎技术,基于关键字的查询信息很难定位到需求,即使能定位也会有很大的信息冗余,效率低下。
从需求个性角度来看,用户需要的是更加符合个人偏好的查询结果,特别是对于潜层需求,前提是必须对不同类别的用户购买行为有所了解,而不是单一标准的产品分类特征。
从衍生需求的模糊性来看,在很多情况下,购买是一种衍生行为,用户曾经购买了一个产品,由于消费此产品而产生了新的需求。衍生需求一般是内生的,用户很难事先知道衍生需求的名称,需要通过对同类产品,相关需求,以及其他人购买情况等提示信息才能明确。
例如我们以亚马逊图书为例来形象地了解一下推荐系统。

如图所示,这是哈利波特的第一部。
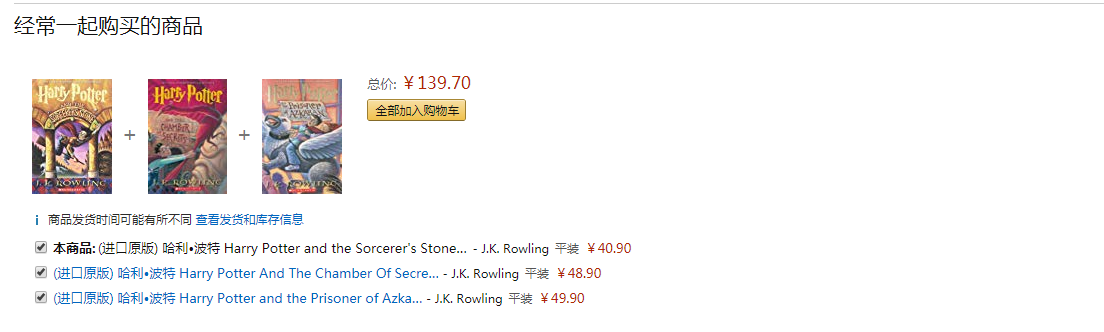
在这张图中,亚马逊的推荐系统将多部哈利波特都推荐了出来,以便于用户的 成套购买,这属于打包销售的策略。

在这张图中,亚马逊列出了曾经购买这本书的用户同时也浏览过的产品,作为同类产品和新颖产品发掘。
上例就是一个经典的个性化推荐系统常用界面,也能反映一个推荐系统的主要功能。我们对上面的各张图进行简单分析,它包括以下几个部分组成:



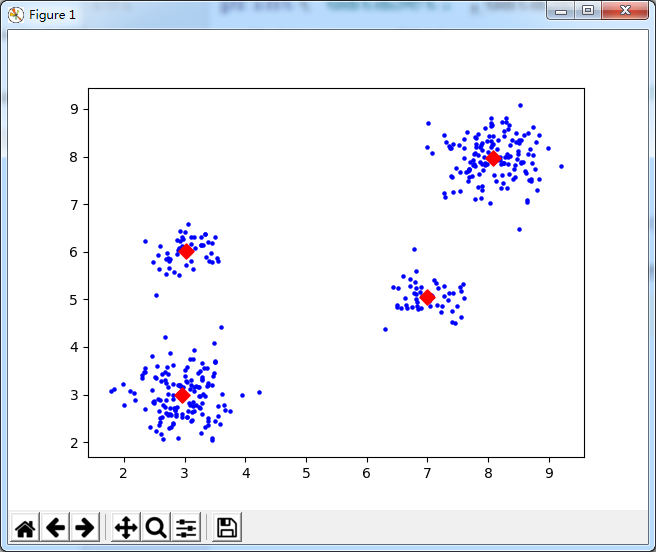
使用Sickit-Learn的KMeans算法对数据进行聚类计算
import numpy as np from sklearn.cluster import KMeans import matplotlib.pyplot as plt #============================================================================================== # -*- coding: utf-8 -*- # Filename : Recommand_lib.py from numpy import * import numpy as np import operator import scipy.spatial.distance as dist # 数据文件转矩阵 # path: 数据文件路径 # delimiter: 文件分隔符 def file2matrix(path, delimiter): recordlist = [] fp = open(path, "rb") # 读取文件内容 content = fp.read().decode() # print("content:",content) fp.close() rowlist = content.splitlines() # 按行转换为一维表 # print("rowlist:",rowlist) # 逐行遍历 # 结果按分隔符分割为行向量 recordlist = [map(eval, row.split(delimiter)) for row in rowlist if row.strip()] testlist = [row.split(delimiter) for row in rowlist if row.strip()] print("testlist:", testlist) testlist2 = (mat(testlist))[:,1:] print("testlist2:",testlist2) # return mat(recordlist) # 返回转换后的矩阵形式 return testlist2 # 随机生成聚类中心 def randCenters(dataSet, k): n = shape(dataSet)[1] clustercents = mat(zeros((k, n))) # 初始化聚类中心矩阵:k*n for col in range(n): mincol = min(dataSet[:, col]); maxcol = max(dataSet[:, col]) # random.rand(k,1): 产生一个0~1之间的随机数向量:k,1表示产生k行1列的随机数 clustercents[:, col] = mat(mincol + float(maxcol - mincol) * random.rand(k, 1)) return clustercents # 欧氏距离 eps = 1.0e-6 def distEclud(vecA, vecB): return linalg.norm(vecA - vecB) + eps # 相关系数 def distCorrcoef(vecA, vecB): return corrcoef(vecA, vecB, rowvar=0)[0][1] # Jaccard距离 def distJaccard(vecA, vecB): temp = mat([array(vecA.tolist()[0]), array(vecB.tolist()[0])]) return dist.pdist(temp, 'jaccard') # 余弦相似度 def cosSim(vecA, vecB): return (dot(vecA, vecB.T) / ((linalg.norm(vecA) * linalg.norm(vecB)) + eps))[0, 0] # 绘制散点图 def drawScatter(plt, mydata, size=20, color='blue', mrkr='o'): plt.scatter(mydata.T[0].tolist(), mydata.T[1].tolist(), s=size, c=color, marker=mrkr) # 根据聚类范围绘制散点图 def color_cluster(dataindx, dataSet, plt, k=4): index = 0 datalen = len(dataindx) for indx in range(datalen): if int(dataindx[indx]) == 0: plt.scatter(dataSet[index, 0], dataSet[index, 1], c='blue', marker='o') elif int(dataindx[indx]) == 1: plt.scatter(dataSet[index, 0], dataSet[index, 1], c='green', marker='o') elif int(dataindx[indx]) == 2: plt.scatter(dataSet[index, 0], dataSet[index, 1], c='red', marker='o') elif int(dataindx[indx]) == 3: plt.scatter(dataSet[index, 0], dataSet[index, 1], c='cyan', marker='o') index += 1 # KMeans 主函数 def kMeans(dataSet, k, distMeas=distEclud, createCent=randCenters): m = shape(dataSet)[0] clusterAssment = mat(zeros((m, 2))) centroids = createCent(dataSet, k) clusterChanged = True while clusterChanged: clusterChanged = False for i in range(m): distlist = [distMeas(centroids[j, :], dataSet[i, :]) for j in range(k)] minDist = min(distlist) minIndex = distlist.index(minDist) if clusterAssment[i, 0] != minIndex: clusterChanged = True clusterAssment[i, :] = minIndex, minDist ** 2 for cent in range(k): # recalculate centroids ptsInClust = dataSet[nonzero(clusterAssment[:, 0].A == cent)[0]] centroids[cent, :] = mean(ptsInClust, axis=0) return centroids, clusterAssment def biKmeans(dataSet, k, distMeas=distEclud): m = shape(dataSet)[0] clusterAssment = mat(zeros((m, 2))) centroid0 = mean(dataSet, axis=0).tolist()[0] centList = [centroid0] # create a list with one centroid for j in range(m): # calc initial Error clusterAssment[j, 1] = distMeas(mat(centroid0), dataSet[j, :]) ** 2 while (len(centList) < k): lowestSSE = inf for i in range(len(centList)): ptsInCurrCluster = dataSet[nonzero(clusterAssment[:, 0].A == i)[0], :] # get the data points currently in cluster i centroidMat, splitClustAss = kMeans(ptsInCurrCluster, 2, distMeas) sseSplit = sum(splitClustAss[:, 1]) # compare the SSE to the currrent minimum sseNotSplit = sum(clusterAssment[nonzero(clusterAssment[:, 0].A != i)[0], 1]) print("sseSplit, and notSplit: ", sseSplit, sseNotSplit) if (sseSplit + sseNotSplit) < lowestSSE: bestCentToSplit = i bestNewCents = centroidMat bestClustAss = splitClustAss.copy() lowestSSE = sseSplit + sseNotSplit bestClustAss[nonzero(bestClustAss[:, 0].A == 1)[0], 0] = len(centList) # change 1 to 3,4, or whatever bestClustAss[nonzero(bestClustAss[:, 0].A == 0)[0], 0] = bestCentToSplit print('the bestCentToSplit is: ', bestCentToSplit) print('the len of bestClustAss is: ', len(bestClustAss)) centList[bestCentToSplit] = bestNewCents[0, :].tolist()[0] # replace a centroid with two best centroids centList.append(bestNewCents[1, :].tolist()[0]) clusterAssment[nonzero(clusterAssment[:, 0].A == bestCentToSplit)[0], :] = bestClustAss # reassign new clusters, and SSE return mat(centList), clusterAssment # dataSet 训练集 # testVect 测试集 # r=3 取前r个近似值 # rank=1,结果排序 # distCalc 相似度计算函数 def recommand(dataSet, testVect, r=3, rank=1, distCalc=cosSim): m, n = shape(dataSet) limit = min(m, n) if r > limit: r = limit U, S, VT = linalg.svd(dataSet.T) # svd分解 V = VT.T Ur = U[:, :r] # 取前r个U,S,V值 Sr = diag(S)[:r, :r] Vr = V[:, :r] testresult = testVect * Ur * linalg.inv(Sr) # 计算User E的坐标值 # 计算测试集与训练集每个记录的相似度 resultarray = array([distCalc(testresult, vi) for vi in Vr]) descindx = argsort(-resultarray)[:rank] # 排序结果--降序 return descindx, resultarray[descindx] # 排序后的索引和值 def kNN(testdata, trainSet, listClasses, k): dataSetSize = trainSet.shape[0] distances = array(zeros(dataSetSize)) for indx in range(dataSetSize): distances[indx] = cosSim(testdata, trainSet[indx]) sortedDistIndicies = argsort(-distances) classCount = {} for i in range(k): # i = 0~(k-1) voteIlabel = listClasses[sortedDistIndicies[i]] classCount[voteIlabel] = classCount.get(voteIlabel, 0) + 1 sortedClassCount = sorted(classCount.iteritems(), key=operator.itemgetter(1), reverse=True) return sortedClassCount[0][0] #============================================================================================== k = 4 dataSet = file2matrix(r"C:UsersAdministratorDesktop estdata4k2_far.txt"," ") print("dataSet:",dataSet) #执行KMeans算法 kmeans = KMeans(init='k-means++',n_clusters=4) kmeans.fit(dataSet) print("--------bbb") drawScatter(plt,dataSet,size=20,color='b',mrkr='.') drawScatter(plt,kmeans.cluster_centers_,size=60,color='red',mrkr='D') plt.show()
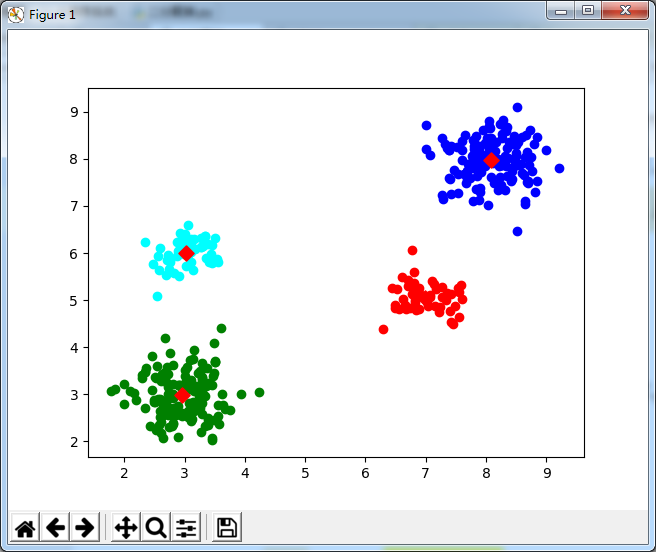
2. 二分聚类python实现
import numpy as np from sklearn.cluster import KMeans import matplotlib.pyplot as plt #============================================================================================== # -*- coding: utf-8 -*- # Filename : Recommand_lib.py from numpy import * import numpy as np import operator import scipy.spatial.distance as dist # 数据文件转矩阵 # path: 数据文件路径 # delimiter: 文件分隔符 def file2matrix(path, delimiter): recordlist = [] fp = open(path, "rb") # 读取文件内容 content = fp.read().decode() # print("content:",content) fp.close() rowlist = content.splitlines() # 按行转换为一维表 # print("rowlist:",rowlist) # 逐行遍历 # 结果按分隔符分割为行向量 recordlist = [map(eval, row.split(delimiter)) for row in rowlist if row.strip()] testlist = [row.split(delimiter) for row in rowlist if row.strip()] # print("testlist:", testlist) testlist2 = (mat(testlist))[:,1:] # print("testlist2:",testlist2) # return mat(recordlist) # 返回转换后的矩阵形式 return testlist2 # 随机生成聚类中心 def randCenters(dataSet, k): n = shape(dataSet)[1] clustercents = mat(zeros((k, n))) # 初始化聚类中心矩阵:k*n for col in range(n): mincol = min(dataSet[:, col]); maxcol = max(dataSet[:, col]) # random.rand(k,1): 产生一个0~1之间的随机数向量:k,1表示产生k行1列的随机数 clustercents[:, col] = mat(mincol + float(maxcol - mincol) * random.rand(k, 1)) return clustercents # 欧氏距离 eps = 1.0e-6 def distEclud(vecA, vecB): return linalg.norm(vecA - vecB) + eps # 相关系数 def distCorrcoef(vecA, vecB): return corrcoef(vecA, vecB, rowvar=0)[0][1] # Jaccard距离 def distJaccard(vecA, vecB): temp = mat([array(vecA.tolist()[0]), array(vecB.tolist()[0])]) return dist.pdist(temp, 'jaccard') # 余弦相似度 def cosSim(vecA, vecB): return (dot(vecA, vecB.T) / ((linalg.norm(vecA) * linalg.norm(vecB)) + eps))[0, 0] # 绘制散点图 def drawScatter(plt, mydata, size=20, color='blue', mrkr='o'): plt.scatter(mydata.T[0].tolist(), mydata.T[1].tolist(), s=size, c=color, marker=mrkr) # 根据聚类范围绘制散点图 def color_cluster(dataindx, dataSet, plt, k=4): index = 0 datalen = len(dataindx) for indx in range(datalen): if int(dataindx[indx]) == 0: plt.scatter(dataSet[index, 0], dataSet[index, 1], c='blue', marker='o') elif int(dataindx[indx]) == 1: plt.scatter(dataSet[index, 0], dataSet[index, 1], c='green', marker='o') elif int(dataindx[indx]) == 2: plt.scatter(dataSet[index, 0], dataSet[index, 1], c='red', marker='o') elif int(dataindx[indx]) == 3: plt.scatter(dataSet[index, 0], dataSet[index, 1], c='cyan', marker='o') index += 1 # KMeans 主函数 def kMeans(dataSet, k, distMeas=distEclud, createCent=randCenters): m = shape(dataSet)[0] clusterAssment = mat(zeros((m, 2))) centroids = createCent(dataSet, k) clusterChanged = True while clusterChanged: clusterChanged = False for i in range(m): distlist = [distMeas(centroids[j, :], dataSet[i, :]) for j in range(k)] minDist = min(distlist) minIndex = distlist.index(minDist) if clusterAssment[i, 0] != minIndex: clusterChanged = True clusterAssment[i, :] = minIndex, minDist ** 2 for cent in range(k): # recalculate centroids ptsInClust = dataSet[nonzero(clusterAssment[:, 0].A == cent)[0]] centroids[cent, :] = mean(ptsInClust, axis=0) return centroids, clusterAssment def biKmeans(dataSet, k, distMeas=distEclud): m = shape(dataSet)[0] clusterAssment = mat(zeros((m, 2))) centroid0 = mean(dataSet, axis=0).tolist()[0] centList = [centroid0] # create a list with one centroid for j in range(m): # calc initial Error clusterAssment[j, 1] = distMeas(mat(centroid0), dataSet[j, :]) ** 2 while (len(centList) < k): lowestSSE = inf for i in range(len(centList)): ptsInCurrCluster = dataSet[nonzero(clusterAssment[:, 0].A == i)[0], :] # get the data points currently in cluster i centroidMat, splitClustAss = kMeans(ptsInCurrCluster, 2, distMeas) sseSplit = sum(splitClustAss[:, 1]) # compare the SSE to the currrent minimum sseNotSplit = sum(clusterAssment[nonzero(clusterAssment[:, 0].A != i)[0], 1]) print("sseSplit, and notSplit: ", sseSplit, sseNotSplit) if (sseSplit + sseNotSplit) < lowestSSE: bestCentToSplit = i bestNewCents = centroidMat bestClustAss = splitClustAss.copy() lowestSSE = sseSplit + sseNotSplit bestClustAss[nonzero(bestClustAss[:, 0].A == 1)[0], 0] = len(centList) # change 1 to 3,4, or whatever bestClustAss[nonzero(bestClustAss[:, 0].A == 0)[0], 0] = bestCentToSplit print('the bestCentToSplit is: ', bestCentToSplit) print('the len of bestClustAss is: ', len(bestClustAss)) centList[bestCentToSplit] = bestNewCents[0, :].tolist()[0] # replace a centroid with two best centroids centList.append(bestNewCents[1, :].tolist()[0]) clusterAssment[nonzero(clusterAssment[:, 0].A == bestCentToSplit)[0], :] = bestClustAss # reassign new clusters, and SSE return mat(centList), clusterAssment # dataSet 训练集 # testVect 测试集 # r=3 取前r个近似值 # rank=1,结果排序 # distCalc 相似度计算函数 def recommand(dataSet, testVect, r=3, rank=1, distCalc=cosSim): m, n = shape(dataSet) limit = min(m, n) if r > limit: r = limit U, S, VT = linalg.svd(dataSet.T) # svd分解 V = VT.T Ur = U[:, :r] # 取前r个U,S,V值 Sr = diag(S)[:r, :r] Vr = V[:, :r] testresult = testVect * Ur * linalg.inv(Sr) # 计算User E的坐标值 # 计算测试集与训练集每个记录的相似度 resultarray = array([distCalc(testresult, vi) for vi in Vr]) descindx = argsort(-resultarray)[:rank] # 排序结果--降序 return descindx, resultarray[descindx] # 排序后的索引和值 def kNN(testdata, trainSet, listClasses, k): dataSetSize = trainSet.shape[0] distances = array(zeros(dataSetSize)) for indx in range(dataSetSize): distances[indx] = cosSim(testdata, trainSet[indx]) sortedDistIndicies = argsort(-distances) classCount = {} for i in range(k): # i = 0~(k-1) voteIlabel = listClasses[sortedDistIndicies[i]] classCount[voteIlabel] = classCount.get(voteIlabel, 0) + 1 sortedClassCount = sorted(classCount.iteritems(), key=operator.itemgetter(1), reverse=True) return sortedClassCount[0][0] #============================================================================================== #导入所需的库,倒入肉数据集,指定聚类中心k k = 4 #分类数 #从文件构建的数据集 dataSet = double(file2matrix(r"C:UsersAdministratorDesktop estdata4k2_far.txt"," ")) m = shape(dataSet)[0] centroid0 = np.mean(dataSet,axis=0) #初始化第一个聚类中心,每一列的均值 # print(centroid0) centList = [centroid0] #把均值聚类中心加入中心表中 # print(centList) ClustDist = mat(zeros((m,2)))#初始化聚类距离表,距离方差 # print(ClustDist) for j in range(m): ClustDist[j,1] = distEclud(centroid0,dataSet[j,:])**2 # 依次生成k个聚类中心 while (len(centList) < k): lowestSSE = inf # 初始化最小误差平方和。核心参数,这个值越小就说明聚类的效果越好。 # 遍历cenList的每个向量 # ----1. 使用ClustDist计算lowestSSE,以此确定:bestCentToSplit、bestNewCents、bestClustAss----# for i in range(len(centList)): ptsInCurrCluster = dataSet[nonzero(ClustDist[:, 0].A == i)[0], :] # 应用标准kMeans算法(k=2),将ptsInCurrCluster划分出两个聚类中心,以及对应的聚类距离表 centroidMat, splitClustAss = kMeans(ptsInCurrCluster, 2) # 计算splitClustAss的距离平方和 sseSplit = sum(splitClustAss[:, 1]) # 计算ClustDist[ClustDist第1列!=i的距离平方和 sseNotSplit = sum(ClustDist[nonzero(ClustDist[:, 0].A != i)[0], 1]) if (sseSplit + sseNotSplit) < lowestSSE: # 算法公式: lowestSSE = sseSplit + sseNotSplit bestCentToSplit = i # 确定聚类中心的最优分隔点 bestNewCents = centroidMat # 用新的聚类中心更新最优聚类中心 bestClustAss = splitClustAss.copy() # 深拷贝聚类距离表为最优聚类距离表 lowestSSE = sseSplit + sseNotSplit # 更新lowestSSE # 回到外循环 # ----2. 计算新的ClustDist----# # 计算bestClustAss 分了两部分: # 第一部分为bestClustAss[bIndx0,0]赋值为聚类中心的索引 bestClustAss[nonzero(bestClustAss[:, 0].A == 1)[0], 0] = len(centList) # 第二部分 用最优分隔点的指定聚类中心索引 bestClustAss[nonzero(bestClustAss[:, 0].A == 0)[0], 0] = bestCentToSplit # 以上为计算bestClustAss # 更新ClustDist对应最优分隔点下标的距离,使距离值等于最优聚类距离对应的值 # 以上为计算ClustDist # ----3. 用最优分隔点来重构聚类中心----# # 覆盖: bestNewCents[0,:].tolist()[0]附加到原有聚类中心的bestCentToSplit位置 # 增加: 聚类中心增加一个新的bestNewCents[1,:].tolist()[0]向量 centList[bestCentToSplit] = bestNewCents[0, :].tolist()[0] centList.append(bestNewCents[1, :].tolist()[0]) ClustDist[nonzero(ClustDist[:, 0].A == bestCentToSplit)[0], :] = bestClustAss # 以上为计算centList color_cluster(ClustDist[:,0:1],dataSet,plt) print("cenList:",mat(centList)) # print "ClustDist:", ClustDist # 绘制聚类中心图形 drawScatter(plt,mat(centList),size=60,color='red',mrkr='D') plt.show()

3.奇异值分解
from numpy import * import numpy as np A = mat([[5,5,3,0,5,5],[5,0,4,0,4,4],[0,3,0,5,4,5],[5,4,3,3,5,5]]) #手工分解求矩阵的svd print("A:") print(A) U = A*A.T print("A*A.T:") print(U) #求U的特征值和特征向量 lamd,hU = linalg.eig(U) VT = A.T*A #求VT的特征值和特征向量 eV,hVT = linalg.eig(VT) hV = hVT.T print("hU:") print(hU) print("hV:") print(hV) sigma = sqrt(lamd) print("sigma:",sigma) #使用函数直接计算 Sigma = np.zeros([shape(A)[0],shape(A)[1]]) u,s,vt = linalg.svd(A) print(s)