TCGA 数据下载分析利器 —— TCGAbiolinks(二)临床数据下载
获取临床信息
TCGAbiolinks 还提供了一些用于查询、下载和解析临床数据的函数,GDC 数据库中的包含多种不同的临床信息,主要包括:
indexed clinical: 使用XML文件创建的精炼临床数据XML: 原始临床数据BCR Biotab: 解析XML文件之后的tsv文件
indexed 信息和 XML 原始数据之间的区别主要有两个:
XML包含的信息更多,如放疗、药物信息、预后信息、样本信息等,而indexed只是XML的一个子集indexed数据包含更新后的预后信息,也就是说,如果在前一次随访中患者还活着,而后一次随访发现患者已经死去了,则会更新患者的状态为死亡,而XML文件会重新添加一个条目,用于记录该次随访的信息
还可以获取其他临床信息,如
- 组织切片图像
- 病理报告
1. BCR Biotab
1.1 Clinical
我们获取乳腺癌 BCR Biotab 文件格式的临床信息
query <- GDCquery(
project = "TCGA-BRCA",
data.category = "Clinical",
data.type = "Clinical Supplement",
data.format = "BCR Biotab"
)
GDCdownload(query)
clinical.BCRtab.all <- GDCprepare(query)
查看具体包含的临床信息,有药物、随访、化疗等信息
> names(clinical.BCRtab.all)
[1] "clinical_drug_brca" "clinical_omf_v4.0_brca" "clinical_follow_up_v4.0_brca"
[4] "clinical_follow_up_v1.5_brca" "clinical_follow_up_v4.0_nte_brca" "clinical_patient_brca"
[7] "clinical_radiation_brca" "clinical_nte_brca" "clinical_follow_up_v2.1_brca"
查看患者信息
> patient_info <- clinical.BCRtab.all$clinical_patient_brca
> dim(patient_info)
[1] 1099 112
> patient_info[1:6, 1:6]
# A tibble: 6 x 6
bcr_patient_uuid bcr_patient_barc… form_completion_… prospective_collec… retrospective_colle… birth_days_to
<chr> <chr> <chr> <chr> <chr> <chr>
1 bcr_patient_uuid bcr_patient_barc… form_completion_… tissue_prospective… tissue_retrospectiv… days_to_birth
2 CDE_ID: CDE_ID:2003301 CDE_ID: CDE_ID:3088492 CDE_ID:3088528 CDE_ID:30082…
3 6E7D5EC6-A469-467C-… TCGA-3C-AAAU 2014-1-13 NO YES -20211
4 55262FCB-1B01-4480-… TCGA-3C-AALI 2014-7-28 NO YES -18538
5 427D0648-3F77-4FFC-… TCGA-3C-AALJ 2014-7-28 NO YES -22848
6 C31900A4-5DCD-4022-… TCGA-3C-AALK 2014-7-28 NO YES -19074
如果我们想获取乳腺癌患者的 er 状态,那么可以
> patient_info %>%
+ dplyr::select(starts_with("er"))
# A tibble: 1,099 x 6
er_status_by_ihc er_status_ihc_Per… er_positivity_scale… er_ihc_score er_positivity_scal… er_positivity_m…
<chr> <chr> <chr> <chr> <chr> <chr>
1 breast_carcinoma_… er_level_cell_per… breast_carcinoma_im… immunohistoch… positive_finding_e… er_detection_me…
2 CDE_ID:2957359 CDE_ID:3128341 CDE_ID:3203081 CDE_ID:2230166 CDE_ID:3086851 CDE_ID:69
3 Positive 50-59% [Not Available] [Not Availabl… [Not Available] [Not Available]
4 Positive <10% [Not Available] [Not Availabl… [Not Available] [Not Available]
5 Positive 90-99% [Not Available] [Not Availabl… [Not Available] [Not Available]
6 Positive 70-79% [Not Available] [Not Availabl… [Not Available] [Not Available]
7 Positive 60-69% 3 Point Scale 2+ [Not Available] [Not Available]
8 Positive 70-79% [Not Available] [Not Availabl… [Not Available] [Not Available]
9 Positive 80-89% [Not Available] [Not Availabl… [Not Available] [Not Available]
10 Positive 70-79% 3 Point Scale 2+ [Not Available] [Not Available]
也可以单独获取某一类的信息,如放疗信息,可以设置 file.type 参数
query <- GDCquery(
project = "TCGA-ACC",
data.category = "Clinical",
data.type = "Clinical Supplement",
data.format = "BCR Biotab",
file.type = "radiation"
)
GDCdownload(query)
clinical.BCRtab.radiation <- GDCprepare(query)
> clinical.BCRtab.radiation$clinical_radiation_acc[1:6, 1:6]
# A tibble: 6 x 6
bcr_patient_uuid bcr_patient_barc… bcr_radiation_ba… bcr_radiation_uuid form_completion… radiation_therap…
<chr> <chr> <chr> <chr> <chr> <chr>
1 bcr_patient_uuid bcr_patient_barc… bcr_radiation_ba… bcr_radiation_uuid form_completion… radiation_type
2 CDE_ID: CDE_ID:2003301 CDE_ID: CDE_ID: CDE_ID: CDE_ID:2842944
3 08E0D412-D4D8-4D13-… TCGA-OR-A5J8 TCGA-OR-A5J8-R54… ACC50D7D-5B8F-4187… 2013-12-18 External
4 DAFB9844-1A5C-4AF8-… TCGA-OR-A5JF TCGA-OR-A5JF-R49… 42BFAD52-D76A-4523… 2013-10-3 External
5 A477B55D-A591-482F-… TCGA-OR-A5JK TCGA-OR-A5JK-R49… 588E4140-E8D4-4E6A… 2013-10-3 External
6 FB54458D-C373-46C2-… TCGA-OR-A5JM TCGA-OR-A5JM-R49… E1ECA707-21FF-43DD… 2013-10-3 Internal
1.2 Biospecimen
获取采样信息
query.biospecimen <- GDCquery(
project = "TCGA-BRCA",
data.category = "Biospecimen",
data.type = "Biospecimen Supplement",
data.format = "BCR Biotab"
)
GDCdownload(query.biospecimen)
biospecimen.BCRtab.all <- GDCprepare(query.biospecimen)
查看所包含的所有信息种类
> names(biospecimen.BCRtab.all)
[1] "biospecimen_aliquot_brca" "ssf_tumor_samples_brca"
[3] "biospecimen_slide_brca" "biospecimen_shipment_portion_brca"
[5] "biospecimen_analyte_brca" "biospecimen_sample_brca"
[7] "biospecimen_portion_brca" "ssf_normal_controls_brca"
[9] "biospecimen_diagnostic_slides_brca" "biospecimen_protocol_brca"
2. indexed
临床 indexed 数据,使用 GDCquery_clinic 函数来获取
clinical <- GDCquery_clinic(project = "TCGA-BRCA", type = "clinical")
> dim(clinical)
[1] 1098 74
> clinical[1:6, 1:6]
submitter_id synchronous_malignancy ajcc_pathologic_stage tumor_stage days_to_diagnosis created_datetime
1 TCGA-E2-A14U No Stage I stage i 0 NA
2 TCGA-E9-A1RC No Stage IIIC stage iiic 0 NA
3 TCGA-D8-A1J9 No Stage IA stage ia 0 NA
4 TCGA-E2-A14P No Stage IIIC stage iiic 0 NA
5 TCGA-A7-A4SD No Stage IIA stage iia 0 NA
6 TCGA-A2-A0CT No Stage IIA stage iia 0 NA
获取其他项目的临床信息
> clinical <- GDCquery_clinic(project = "GENIE-MSK", type = "clinical")
> dim(clinical)
[1] 16824 125
> clinical[1:6, 14:17]
state cog_rhabdomyosarcoma_risk_group primary_gleason_grade morphology
1 released NA NA 8810/3
2 released NA NA 8140/3
3 released NA NA 8140/3
4 released NA NA 9133/3
5 released NA NA 8140/3
6 released NA NA 8441/3
3. XML
处理 XML 格式的临床数据分为两步:
- 使用
GDCquery和GDCDownload来查询和下载Biospecimen或ClinicalXML文件 - 使用
GDCprepare_clinic来解析文件
注意:患者与临床信息是一对多的关系,即一个患者可能会接受多次化疗,因此,只能对单个表进行解析,使用 clinical.info 参数来选择对应的表
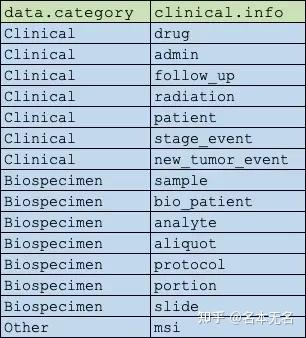
query <- GDCquery(
project = "TCGA-BRCA",
data.category = "Clinical",
file.type = "xml",
barcode = c("TCGA-3C-AAAU", "TCGA-4H-AAAK")
)
GDCdownload(query)
解析患者表
clinical <- GDCprepare_clinic(query, clinical.info = "patient")
> clinical[,1:6]
bcr_patient_barcode additional_studies tumor_tissue_site tumor_tissue_site_other other_dx gender
1 TCGA-3C-AAAU NA Breast NA No FEMALE
2 TCGA-4H-AAAK NA Breast NA No FEMALE
获取用药信息
> clinical.drug <- GDCprepare_clinic(query, clinical.info = "drug")
> clinical.drug[,1:4]
bcr_patient_barcode tx_on_clinical_trial regimen_number bcr_drug_barcode
1 TCGA-3C-AAAU YES NA TCGA-3C-AAAU-D60350
2 TCGA-4H-AAAK NO NA TCGA-4H-AAAK-D68065
3 TCGA-4H-AAAK NO NA TCGA-4H-AAAK-D68067
4 TCGA-4H-AAAK NO NA TCGA-4H-AAAK-D68072
由于这两个患者没有放疗信息,返回了 NULL
> clinical.radiation <- GDCprepare_clinic(query, clinical.info = "radiation")
| | 0%
> clinical.radiation
NULL
4. 其他数据
4.1 MSI 状态
样本的 MSI(微卫星不稳定性)状态是通过 MSI-Mono-Dinucleotide Assay 来检测的,如果样本的状态不明确,会再使用 PCR 进行确认
query <- GDCquery(
project = "TCGA-COAD",
data.category = "Other",
legacy = TRUE,
access = "open",
data.type = "Auxiliary test",
barcode = c("TCGA-AD-A5EJ", "TCGA-DM-A0X9")
)
GDCdownload(query)
msi_results <- GDCprepare_clinic(query, "msi")
> msi_results[,c(1, 3)]
bcr_patient_barcode mononucleotide_and_dinucleotide_marker_panel_analysis_status
1 TCGA-AD-A5EJ MSI-H
2 TCGA-DM-A0X9 MSS
4.2 组织切片图像(SVS 格式)
# legacy database
query.legacy <- GDCquery(
project = "TCGA-COAD",
data.category = "Clinical",
data.type = "Tissue slide image",
legacy = TRUE,
barcode = c("TCGA-RU-A8FL", "TCGA-AA-3972")
)
# harmonized database
query.harmonized <- GDCquery(
project = "TCGA-OV",
data.category = "Biospecimen",
data.type = 'Slide Image'
)
legacy 数据
> getResults(query.legacy)[,1:4]
id data_format access cases
688 530083be-6bdf-49e8-85dc-3c3ee5d5dcd5 SVS open TCGA-RU-A8FL
315 0d4a3c6c-0ab2-44d2-9b08-90f5ea84555f SVS open TCGA-AA-3972
320 4966c8e3-37fd-4296-8a8a-216def9ec311 SVS open TCGA-AA-3972
harmonized 数据
> getResults(query.harmonized)[1:6,1:4]
id data_format cases access
1 4f0fc759-1f71-4d01-a602-0e07abd35120 SVS TCGA-13-1499 open
2 812b0b13-58df-42ee-be06-04e56720a3cc SVS TCGA-61-1724 open
3 f2521543-5a5d-4f24-bd77-b084ec4e035f SVS TCGA-04-1638 open
4 c028facc-d2be-4acb-9ea2-05987de0199f SVS TCGA-04-1351 open
5 f39eac18-1a69-4cb1-b962-d1d3135703f9 SVS TCGA-23-2643 open
6 ea326c8e-0500-4fb9-a0f3-607206d7fdd1 SVS TCGA-61-1728 open
4.3 诊断切片(SVS 格式)
query.harmonized <- GDCquery(
project = "TCGA-COAD",
data.category = "Biospecimen",
data.type = "Slide Image",
experimental.strategy = "Diagnostic Slide",
barcode = c("TCGA-RU-A8FL", "TCGA-AA-3972")
)
> getResults(query.harmonized)[,1:4]
id data_format access cases
226 b339b9d1-af19-46e3-94cf-eb21c391da0e SVS open TCGA-AA-3972
5. Legacy 临床数据
Legacy 数据库中包含如下临床数据类型:
Biospecimen data(Biotab)Tissue slide image(SVS)Clinical Supplement(XML)Pathology report(PDF)Clinical data(Biotab)
5.1 病理报告
query.legacy <- GDCquery(
project = "TCGA-COAD",
data.category = "Clinical",
data.type = "Pathology report",
legacy = TRUE,
barcode = c("TCGA-RU-A8FL", "TCGA-AA-3972")
)
> getResults(query.legacy)[, 1:4]
id data_format access cases
7 a4753077-2bd3-4301-8424-b7575c8ccd66 PDF open TCGA-RU-A8FL
365 b77a41e9-cf0d-4b94-9576-09e91b6d8f61 PDF open TCGA-AA-3972
5.2 组织切片
query <- GDCquery(
project = "TCGA-COAD",
data.category = "Clinical",
data.type = "Tissue slide image",
legacy = TRUE,
barcode = c("TCGA-RU-A8FL", "TCGA-AA-3972")
)
> getResults(query)[, 1:4]
id data_format access cases
688 530083be-6bdf-49e8-85dc-3c3ee5d5dcd5 SVS open TCGA-RU-A8FL
315 0d4a3c6c-0ab2-44d2-9b08-90f5ea84555f SVS open TCGA-AA-3972
320 4966c8e3-37fd-4296-8a8a-216def9ec311 SVS open TCGA-AA-3972
5.3 临床信息
query <- GDCquery(
project = "TCGA-COAD",
data.category = "Clinical",
data.type = "Clinical data",
legacy = TRUE,
file.type = "txt"
)
GDCdownload(query)
clinical.biotab <- GDCprepare(query)
> getResults(query)[, 1:4]
id data_format access cases
26 36b48c2d-f45d-4995-bc2d-931f5c190919 Biotab open <NA>
36 b58b5947-d2b6-4cc7-9eff-cc0083d5bf4b Biotab open <NA>
38 0415ffe2-a98d-40b9-ac60-6753fce56c7b Biotab open <NA>
45 b0e4e0aa-2398-4a31-b205-656da0100c06 Biotab open <NA>
49 2cf76252-d084-4653-969e-7264332b1bdb Biotab open <NA>
57 1a624a8a-fb61-43c9-8f6f-83528466029e Biotab open <NA>
66 eba36073-8181-4415-81d0-e2791723f571 Biotab open <NA>
> names(clinical.biotab)
[1] "clinical_radiation_coad" "clinical_patient_coad" "clinical_drug_coad"
[4] "clinical_follow_up_v1.0_coad" "clinical_nte_coad" "clinical_follow_up_v1.0_nte_coad"
[7] "clinical_omf_v4.0_coad"
5.4 临床补充信息
query <- GDCquery(
project = "TCGA-COAD",
data.category = "Clinical",
data.type = "Clinical Supplement",
legacy = TRUE,
barcode = c("TCGA-RU-A8FL", "TCGA-AA-3972")
)
> getResults(query)[, 1:4]
id data_format access cases
303 3c5a4713-6855-42d4-aed6-3129bfe80c58 BCR XML open TCGA-RU-A8FL
99 c76af5df-aab0-47a0-a543-77668be3f0c7 BCR XML open TCGA-AA-3972
6. 过滤函数
还有一些函数用过筛选临床样本,例如
TCGAquery_SampleTypes:
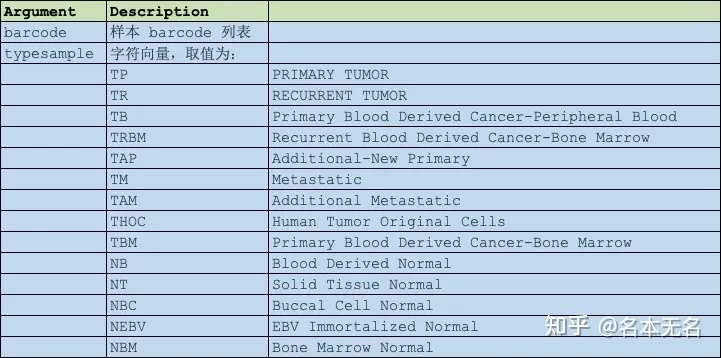
bar <- c("TCGA-G9-6378-02A-11R-1789-07", "TCGA-CH-5767-04A-11R-1789-07",
"TCGA-G9-6332-60A-11R-1789-07", "TCGA-G9-6336-01A-11R-1789-07",
"TCGA-G9-6336-11A-11R-1789-07", "TCGA-G9-7336-11A-11R-1789-07",
"TCGA-G9-7336-04A-11R-1789-07", "TCGA-G9-7336-14A-11R-1789-07",
"TCGA-G9-7036-04A-11R-1789-07", "TCGA-G9-7036-02A-11R-1789-07",
"TCGA-G9-7036-11A-11R-1789-07", "TCGA-G9-7036-03A-11R-1789-07",
"TCGA-G9-7036-10A-11R-1789-07", "TCGA-BH-A1ES-10A-11R-1789-07",
"TCGA-BH-A1F0-10A-11R-1789-07", "TCGA-BH-A0BZ-02A-11R-1789-07",
"TCGA-B6-A0WY-04A-11R-1789-07", "TCGA-BH-A1FG-04A-11R-1789-08",
"TCGA-D8-A1JS-04A-11R-2089-08", "TCGA-AN-A0FN-11A-11R-8789-08",
"TCGA-AR-A2LQ-12A-11R-8799-08", "TCGA-AR-A2LH-03A-11R-1789-07",
"TCGA-BH-A1F8-04A-11R-5789-07", "TCGA-AR-A24T-04A-55R-1789-07",
"TCGA-AO-A0J5-05A-11R-1789-07", "TCGA-BH-A0B4-11A-12R-1789-07",
"TCGA-B6-A1KN-60A-13R-1789-07", "TCGA-AO-A0J5-01A-11R-1789-07",
"TCGA-AO-A0J5-01A-11R-1789-07", "TCGA-G9-6336-11A-11R-1789-07",
"TCGA-G9-6380-11A-11R-1789-07", "TCGA-G9-6380-01A-11R-1789-07",
"TCGA-G9-6340-01A-11R-1789-07", "TCGA-G9-6340-11A-11R-1789-07")
S <- TCGAquery_SampleTypes(bar,"TP")
S2 <- TCGAquery_SampleTypes(bar,"NB")
# 返回 TP 或 NB 类型的样本
SS <- TCGAquery_SampleTypes(bar,c("TP","NB"))
> S
[1] "TCGA-G9-6336-01A-11R-1789-07" "TCGA-AO-A0J5-01A-11R-1789-07" "TCGA-G9-6380-01A-11R-1789-07"
[4] "TCGA-G9-6340-01A-11R-1789-07"
> S2
[1] "TCGA-G9-7036-10A-11R-1789-07" "TCGA-BH-A1ES-10A-11R-1789-07" "TCGA-BH-A1F0-10A-11R-1789-07"
> SS
[1] "TCGA-G9-6336-01A-11R-1789-07" "TCGA-AO-A0J5-01A-11R-1789-07" "TCGA-G9-6380-01A-11R-1789-07"
[4] "TCGA-G9-6340-01A-11R-1789-07" "TCGA-G9-7036-10A-11R-1789-07" "TCGA-BH-A1ES-10A-11R-1789-07"
[7] "TCGA-BH-A1F0-10A-11R-1789-07"
TCGAquery_MatchedCoupledSampleTypes
> SSS <- TCGAquery_MatchedCoupledSampleTypes(bar,c("NT","TP"))
> # 返回同时包含 NT 和 TP 样本的患者 barcode
> SSS
[1] "TCGA-G9-6336-11A-11R-1789-07" "TCGA-G9-6380-11A-11R-1789-07" "TCGA-G9-6340-11A-11R-1789-07"
[4] "TCGA-G9-6336-01A-11R-1789-07" "TCGA-G9-6380-01A-11R-1789-07" "TCGA-G9-6340-01A-11R-1789-07"
7. 下载所有临床数据
library(tidyverse)
getclinical <- function(proj, filename){
message(proj)
while(1){
result = tryCatch({
query <- GDCquery(project = proj, data.category = "Clinical",file.type = "xml")
GDCdownload(query)
clinical <- GDCprepare_clinic(query, clinical.info = "patient")
for(i in c("admin","radiation","follow_up","drug", "stage_event", "new_tumor_event")){
message(i)
aux <- GDCprepare_clinic(query, clinical.info = i)
if(is.null(aux) || nrow(aux) == 0) next
# add suffix manually if it already exists
replicated <- which(grep("bcr_patient_barcode", colnames(aux), value = T,invert = T) %in% colnames(clinical))
colnames(aux)[replicated] <- paste0(colnames(aux)[replicated], ".", i)
if(!is.null(aux))
clinical <- full_join(clinical, aux, by = "bcr_patient_barcode")
}
# 保存为 excel 文件中的一个 sheet
xlsx::write.xlsx(clinical, file = filename, sheetName = proj)
# return(clinical)
return(TRUE)
}, error = function(e) {
message(paste0("Error clinical: ", proj))
})
message("try again!")
}
}
filename <- "~/Downloads/TCGA_clinical.xlsx"
clinical <- TCGAbiolinks:::getGDCprojects()$project_id %>%
regexPipes::grep("TCGA",value=T) %>% sort %>%
map_chr(getclinical, filename)
获取代码:https://github.com/dxsbiocc/learn/blob/main/R/TCGA/download_clinical.R
- END -
发布于 07-04 16:02