斯坦福deep learning教程中的自稀疏编码器的练习,主要是参考了 http://www.cnblogs.com/tornadomeet/archive/2013/03/20/2970724.html,没有参考肯定编不出来。。。Σ( ° △ °|||)︴ 也当自己理解了一下
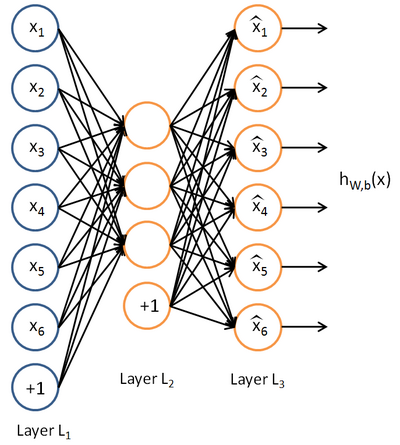
这里的自稀疏编码器,练习上规定是64个输入节点,25个隐藏层节点(我实验中只有20个),输出层也是64个节点,一共有10000个训练样本
具体步骤:
首先在页面上下载sparseae_exercise.zip
Step 1:构建训练集
要求在10张图片(图片数据存储在IMAGES中)中随机的选取一张图片,在再这张图片中随机的选取10000个像素点,最终构建一个64*10000的像素矩阵。从一张图片中选取10000个像素点的好处是,只有copy一次IMAGES,速度更快,但是要注意每张图片的像素是512*512的,所以随机选取像素点最好是分行和列各选取100,最终组合成100*100,这样不容易导致越界。验证step 1可以运行train.m中的第一步,结果图如下:

(只展示了200个sample,所以有4个缺口)
需要自行编写sampleIMAGES中的部分code
function patches = sampleIMAGES() % sampleIMAGES % Returns 10000 patches for training load IMAGES; % load images from disk patchsize = 8; % we'll use 8x8 patches numpatches = 10000; % Initialize patches with zeros. Your code will fill in this matrix--one % column per patch, 10000 columns. patches = zeros(patchsize*patchsize, numpatches); %% ---------- YOUR CODE HERE -------------------------------------- % Instructions: Fill in the variable called "patches" using data % from IMAGES. % % IMAGES is a 3D array containing 10 images % For instance, IMAGES(:,:,6) is a 512x512 array containing the 6th image, % and you can type "imagesc(IMAGES(:,:,6)), colormap gray;" to visualize % it. (The contrast on these images look a bit off because they have % been preprocessed using using "whitening." See the lecture notes for % more details.) As a second example, IMAGES(21:30,21:30,1) is an image % patch corresponding to the pixels in the block (21,21) to (30,30) of % Image 1 imageNum = randi([1,10]); %随机的选择一张图片 [rowNum colNum] = size(IMAGES(:,:,imageNum)); xPos = randperm(rowNum-patchsize+1,100); yPos = randperm(colNum-patchsize+1,100); for ii = 1:100 %在图片中选取100*100个像素点 for jj = 1:100 patchNum = (ii-1)*100 + jj; patches(:,patchNum) = reshape(IMAGES(xPos(ii):xPos(ii)+7,yPos(jj):yPos(jj)+7,... imageNum),64,1); end end %% --------------------------------------------------------------- % For the autoencoder to work well we need to normalize the data % Specifically, since the output of the network is bounded between [0,1] % (due to the sigmoid activation function), we have to make sure % the range of pixel values is also bounded between [0,1] patches = normalizeData(patches); end %% --------------------------------------------------------------- function patches = normalizeData(patches) % Squash data to [0.1, 0.9] since we use sigmoid as the activation % function in the output layer % Remove DC (mean of images). patches = bsxfun(@minus, patches, mean(patches)); % Truncate to +/-3 standard deviations and scale to -1 to 1 pstd = 3 * std(patches(:)); patches = max(min(patches, pstd), -pstd) / pstd; % Rescale from [-1,1] to [0.1,0.9] patches = (patches + 1) * 0.4 + 0.1; end
Step 2:求解自稀疏编码器的参数
这一步就是要运用BP算法求解NN中各层的W,b(W1,W2,b1,b2)参数。 Backpropagation Algorithm算法在教程的第二节中有介绍,但要注意的是自稀疏编码器的误差函数除了有参数的正则化项,还有稀疏性规则项,BP算法推导公式中要加上,这里需要自行编写sparseAutoencoderCost.m
function [cost,grad] = sparseAutoencoderCost(theta, visibleSize, hiddenSize, ... lambda, sparsityParam, beta, data) % visibleSize: the number of input units (probably 64) % hiddenSize: the number of hidden units (probably 25) % lambda: weight decay parameter % sparsityParam: The desired average activation for the hidden units (denoted in the lecture % notes by the greek alphabet rho, which looks like a lower-case "p"). % beta: weight of sparsity penalty term % data: Our 64x10000 matrix containing the training data. So, data(:,i) is the i-th training example. % The input theta is a vector (because minFunc expects the parameters to be a vector). % We first convert theta to the (W1, W2, b1, b2) matrix/vector format, so that this % follows the notation convention of the lecture notes. W1 = reshape(theta(1:hiddenSize*visibleSize), hiddenSize, visibleSize); W2 = reshape(theta(hiddenSize*visibleSize+1:2*hiddenSize*visibleSize), visibleSize, hiddenSize); b1 = theta(2*hiddenSize*visibleSize+1:2*hiddenSize*visibleSize+hiddenSize); b2 = theta(2*hiddenSize*visibleSize+hiddenSize+1:end); % Cost and gradient variables (your code needs to compute these values). % Here, we initialize them to zeros. cost = 0; W1grad = zeros(size(W1)); W2grad = zeros(size(W2)); b1grad = zeros(size(b1)); b2grad = zeros(size(b2)); %% ---------- YOUR CODE HERE -------------------------------------- % Instructions: Compute the cost/optimization objective J_sparse(W,b) for the Sparse Autoencoder, % and the corresponding gradients W1grad, W2grad, b1grad, b2grad. % % W1grad, W2grad, b1grad and b2grad should be computed using backpropagation. % Note that W1grad has the same dimensions as W1, b1grad has the same dimensions % as b1, etc. Your code should set W1grad to be the partial derivative of J_sparse(W,b) with % respect to W1. I.e., W1grad(i,j) should be the partial derivative of J_sparse(W,b) % with respect to the input parameter W1(i,j). Thus, W1grad should be equal to the term % [(1/m) Delta W^{(1)} + lambda W^{(1)}] in the last block of pseudo-code in Section 2.2 % of the lecture notes (and similarly for W2grad, b1grad, b2grad). % % Stated differently, if we were using batch gradient descent to optimize the parameters, % the gradient descent update to W1 would be W1 := W1 - alpha * W1grad, and similarly for W2, b1, b2. % Jcost = 0;%直接误差 Jweight = 0;%权值惩罚 Jsparse = 0;%稀疏性惩罚 [n m] = size(data);%m为样本的个数,n为样本的特征数 %前向算法计算各神经网络节点的线性组合值和active值 z2 = W1*data+repmat(b1,1,m);%注意这里一定要将b1向量复制扩展成m列的矩阵 a2 = sigmoid(z2); z3 = W2*a2+repmat(b2,1,m); a3 = sigmoid(z3); % 计算预测产生的误差 Jcost = (0.5/m)*sum(sum((a3-data).^2)); %计算权值惩罚项 Jweight = (1/2)*(sum(sum(W1.^2))+sum(sum(W2.^2))); %计算稀释性规则项 rho = (1/m).*sum(a2,2) ;%求出第一个隐含层的平均值向量 Jsparse = sum(sparsityParam.*log(sparsityParam./rho)+ ... (1-sparsityParam).*log((1-sparsityParam)./(1-rho))); %损失函数的总表达式 cost = Jcost+lambda*Jweight+beta*Jsparse; %反向算法求出每个节点的误差值 d3 = -(data-a3).*(a3.*(1-a3)); sterm = beta*(-sparsityParam./rho+(1-sparsityParam)./(1-rho));%因为加入了稀疏规则项,所以 %计算偏导时需要引入该项 d2 = (W2'*d3+repmat(sterm,1,m)).*(a2.*(1-a2)); %计算W1grad W1grad = W1grad+d2*data'; W1grad = (1/m)*W1grad+lambda*W1; %计算W2grad W2grad = W2grad+d3*a2'; W2grad = (1/m).*W2grad+lambda*W2; %计算b1grad b1grad = b1grad+sum(d2,2); b1grad = (1/m)*b1grad;%注意b的偏导是一个向量,所以这里应该把每一行的值累加起来 %计算b2grad b2grad = b2grad+sum(d3,2); b2grad = (1/m)*b2grad; %------------------------------------------------------------------- % After computing the cost and gradient, we will convert the gradients back % to a vector format (suitable for minFunc). Specifically, we will unroll % your gradient matrices into a vector. grad = [W1grad(:) ; W2grad(:) ; b1grad(:) ; b2grad(:)]; end %------------------------------------------------------------------- % Here's an implementation of the sigmoid function, which you may find useful % in your computation of the costs and the gradients. This inputs a (row or % column) vector (say (z1, z2, z3)) and returns (f(z1), f(z2), f(z3)). function sigm = sigmoid(x) % 定义sigmoid函数 sigm = 1 ./ (1 + exp(-x)); end
Step 3:求解的 梯度检验
验证梯度下降是否正确,这个在教程第三节也有介绍,比较简单,在computeNumericalGradient.m中返回梯度检验后的值即可,computeNumericalGradient.m是在checkNumericalGradient.m中调用的,而checkNumericalGradient.m已经给出,不需要我们自己编写。
function numgrad = computeNumericalGradient(J, theta) % numgrad = computeNumericalGradient(J, theta) % theta: a vector of parameters % J: a function that outputs a real-number. Calling y = J(theta) will return the % function value at theta. % Initialize numgrad with zeros numgrad = zeros(size(theta)); %% ---------- YOUR CODE HERE -------------------------------------- % Instructions: % Implement numerical gradient checking, and return the result in numgrad. % (See Section 2.3 of the lecture notes.) % You should write code so that numgrad(i) is (the numerical approximation to) the % partial derivative of J with respect to the i-th input argument, evaluated at theta. % I.e., numgrad(i) should be the (approximately) the partial derivative of J with % respect to theta(i). % % Hint: You will probably want to compute the elements of numgrad one at a time. epsilon = 1e-4; n = size(theta,1); E = eye(n,1); for i = 1:n
E(i) = 1; delta = E*epsilon; numgrad(i) = (J(theta+delta)-J(theta-delta))/(epsilon*2.0);
E(i) = 0; end %% --------------------------------------------------------------- end
Step 4:训练自稀疏编码器
整个训练过程使用的是L-BFGS求解,比教程中介绍的主要介绍批量SGD要快很多,具体原理我也不知道,而且训练过程已经给出,这一段不需要我们自己编写
Step 5:输出可视化结果
训练结束后,输出训练得到的权重矩阵W1,结果同时也会保存在weights.jpg中,这一段也不需要我们编写( 第一次)
结果图如下:

(感觉自己训练出来的这个没有标准的那么明显的线条,看就了还有点类似错误示例的第3个,不过重新仔细看还是有线条感的,可能是因为隐藏层只有20个,训练的也没有25个的彻底)
另外,查了一下内存不足的解决方法,据说在matlab命令行输入pack,可以释放一些内存。但是我觉得还是终究治标不治本,最好的方法还是升级64位操作系统,去添加内存条吧~
剩下的.m文件都不需要我们自己编写(修改隐藏层的节点数在train.m中),不过也顺带附上吧

function [] = checkNumericalGradient() % This code can be used to check your numerical gradient implementation % in computeNumericalGradient.m % It analytically evaluates the gradient of a very simple function called % simpleQuadraticFunction (see below) and compares the result with your numerical % solution. Your numerical gradient implementation is incorrect if % your numerical solution deviates too much from the analytical solution. % Evaluate the function and gradient at x = [4; 10]; (Here, x is a 2d vector.) x = [4; 10]; [value, grad] = simpleQuadraticFunction(x); % Use your code to numerically compute the gradient of simpleQuadraticFunction at x. % (The notation "@simpleQuadraticFunction" denotes a pointer to a function.) numgrad = computeNumericalGradient(@simpleQuadraticFunction, x); % Visually examine the two gradient computations. The two columns % you get should be very similar. disp([numgrad grad]); fprintf('The above two columns you get should be very similar. (Left-Your Numerical Gradient, Right-Analytical Gradient) '); % Evaluate the norm of the difference between two solutions. % If you have a correct implementation, and assuming you used EPSILON = 0.0001 % in computeNumericalGradient.m, then diff below should be 2.1452e-12 diff = norm(numgrad-grad)/norm(numgrad+grad); disp(diff); fprintf('Norm of the difference between numerical and analytical gradient (should be < 1e-9) '); end function [value,grad] = simpleQuadraticFunction(x) % this function accepts a 2D vector as input. % Its outputs are: % value: h(x1, x2) = x1^2 + 3*x1*x2 % grad: A 2x1 vector that gives the partial derivatives of h with respect to x1 and x2 % Note that when we pass @simpleQuadraticFunction(x) to computeNumericalGradients, we're assuming % that computeNumericalGradients will use only the first returned value of this function. value = x(1)^2 + 3*x(1)*x(2); grad = zeros(2, 1); grad(1) = 2*x(1) + 3*x(2); grad(2) = 3*x(1); end

function theta = initializeParameters(hiddenSize, visibleSize) %% Initialize parameters randomly based on layer sizes. r = sqrt(6) / sqrt(hiddenSize+visibleSize+1); % we'll choose weights uniformly from the interval [-r, r] W1 = rand(hiddenSize, visibleSize) * 2 * r - r; W2 = rand(visibleSize, hiddenSize) * 2 * r - r; b1 = zeros(hiddenSize, 1); b2 = zeros(visibleSize, 1); % Convert weights and bias gradients to the vector form. % This step will "unroll" (flatten and concatenate together) all % your parameters into a vector, which can then be used with minFunc. theta = [W1(:) ; W2(:) ; b1(:) ; b2(:)]; end

function [h, array] = display_network(A, opt_normalize, opt_graycolor, cols, opt_colmajor) % This function visualizes filters in matrix A. Each column of A is a % filter. We will reshape each column into a square image and visualizes % on each cell of the visualization panel. % All other parameters are optional, usually you do not need to worry % about it. % opt_normalize: whether we need to normalize the filter so that all of % them can have similar contrast. Default value is true. % opt_graycolor: whether we use gray as the heat map. Default is true. % cols: how many columns are there in the display. Default value is the % squareroot of the number of columns in A. % opt_colmajor: you can switch convention to row major for A. In that % case, each row of A is a filter. Default value is false. warning off all if ~exist('opt_normalize', 'var') || isempty(opt_normalize) opt_normalize= true; end if ~exist('opt_graycolor', 'var') || isempty(opt_graycolor) opt_graycolor= true; end if ~exist('opt_colmajor', 'var') || isempty(opt_colmajor) opt_colmajor = false; end % rescale A = A - mean(A(:)); if opt_graycolor, colormap(gray); end % compute rows, cols [L M]=size(A); sz=sqrt(L); buf=1; if ~exist('cols', 'var') if floor(sqrt(M))^2 ~= M n=ceil(sqrt(M)); while mod(M, n)~=0 && n<1.2*sqrt(M), n=n+1; end m=ceil(M/n); else n=sqrt(M); m=n; end else n = cols; m = ceil(M/n); end array=-ones(buf+m*(sz+buf),buf+n*(sz+buf)); if ~opt_graycolor array = 0.1.* array; end if ~opt_colmajor k=1; for i=1:m for j=1:n if k>M, continue; end clim=max(abs(A(:,k))); if opt_normalize array(buf+(i-1)*(sz+buf)+(1:sz),buf+(j-1)*(sz+buf)+(1:sz))=reshape(A(:,k),sz,sz)/clim; else array(buf+(i-1)*(sz+buf)+(1:sz),buf+(j-1)*(sz+buf)+(1:sz))=reshape(A(:,k),sz,sz)/max(abs(A(:))); end k=k+1; end end else k=1; for j=1:n for i=1:m if k>M, continue; end clim=max(abs(A(:,k))); if opt_normalize array(buf+(i-1)*(sz+buf)+(1:sz),buf+(j-1)*(sz+buf)+(1:sz))=reshape(A(:,k),sz,sz)/clim; else array(buf+(i-1)*(sz+buf)+(1:sz),buf+(j-1)*(sz+buf)+(1:sz))=reshape(A(:,k),sz,sz); end k=k+1; end end end if opt_graycolor h=imagesc(array,'EraseMode','none',[-1 1]); else h=imagesc(array,'EraseMode','none',[-1 1]); end axis image off drawnow; warning on all