megablast 采用贪婪式算法,速度较一般blast快,多用于数据量大且序列相似性较高的情况。
megablast 参数说明
./megablast --help
megablast 2.2.11 arguments:
-d Database [String]
default = nr
-i Query File [File In]
-e Expectation value [Real]
default = 10.0
-m alignment view options:
0 = pairwise,
1 = query-anchored showing identities,
2 = query-anchored no identities,
3 = flat query-anchored, show identities,
4 = flat query-anchored, no identities,
5 = query-anchored no identities and blunt ends,
6 = flat query-anchored, no identities and blunt ends,
7 = XML Blast output,
8 = tabular,
9 tabular with comment lines,
10 ASN, text
11 ASN, binary [Integer]
default = 0
-o BLAST report Output File [File Out] Optional
default = stdout
-F Filter query sequence [String]
default = T
-X X dropoff value for gapped alignment (in bits) [Integer]
default = 20
-I Show GI's in deflines [T/F]
default = F
-q Penalty for a nucleotide mismatch [Integer]
default = -3
-r Reward for a nucleotide match [Integer]
default = 1
-v Number of database sequences to show one-line descriptions for (V) [Intege
r]
default = 500
-b Number of database sequence to show alignments for (B) [Integer]
default = 250
-D Type of output:
0 - alignment endpoints and score,
1 - all ungapped segments endpoints,
2 - traditional BLAST output,
3 - tab-delimited one line format [Integer]
default = 2
-a Number of processors to use [Integer]
default = 1
-O ASN.1 SeqAlign file; must be used in conjunction with -D2 option [File Out
] Optional
-J Believe the query defline [T/F] Optional
default = F
-M Maximal total length of queries for a single search [Integer]
default = 20000000
-W Word size (length of best perfect match) [Integer]
default = 28
-z Effective length of the database (use zero for the real size) [Real]
default = 0
-P Maximal number of positions for a hash value (set to 0 to ignore) [Integer
]
default = 0
-S Query strands to search against database: 3 is both, 1 is top, 2 is bottom
[Integer]
default = 3
-T Produce HTML output [T/F]
default = F
-l Restrict search of database to list of GI's [String] Optional
-G Cost to open a gap (zero invokes default behavior) [Integer]
default = 0
-E Cost to extend a gap (zero invokes default behavior) [Integer]
default = 0
-s Minimal hit score to report (0 for default behavior) [Integer]
default = 0
-Q Masked query output, must be used in conjunction with -D 2 option [File Ou
t] Optional
-f Show full IDs in the output (default - only GIs or accessions) [T/F]
default = F
-U Use lower case filtering of FASTA sequence [T/F] Optional
default = F
-R Report the log information at the end of output [T/F] Optional
default = F
-p Identity percentage cut-off [Real]
default = 0
-L Location on query sequence [String] Optional
-A Multiple Hits window size [Integer]
default = 0
-y X dropoff value for ungapped extension [Integer]
default = 10
-Z X dropoff value for dynamic programming gapped extension [Integer]
default = 50
-t Length of a discontiguous word template (contiguous word if 0) [Integer]
default = 0
-g Generate words for every base of the database (default is every 4th base;
may only be used with discontiguous words) [T/F] Optional
default = F
-n Use non-greedy (dynamic programming) extension for affine gap scores [T/F]
Optional
default = F
-N Type of a discontiguous word template (0 - coding, 1 - optimal, 2 - two si
multaneous [Integer]
default = 0
-H Maximal number of HSPs to save per database sequence (0 = unlimited) [Inte
ger]
default = 0
-V Force use of the legacy BLAST engine [T/F] Optional
default = F
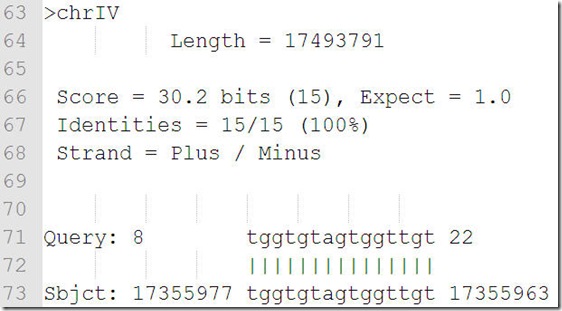
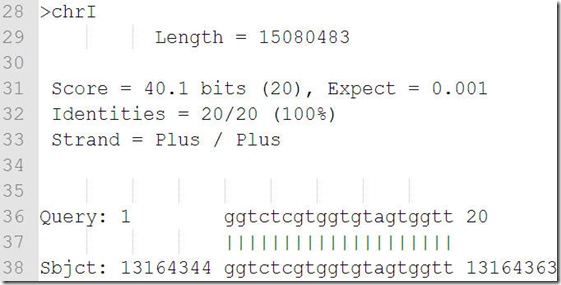
megablast 输出结果
score是打分,打分越高,应该是相似性越高;
expect值越低匹配越好;
identities是一致性,这个参数好像是随机给出的,不能设定;
Strand = Plus / Plus
Strand = Plus / Minus 分别代表匹配在两条不同的链上;
blastn参数:
-db: 指定blast搜索用的数据库
-query:用来查询的输入序列,fasta格式
-out:输出结果文件
-evalue: 设置e值cutoff
-max_target_seqs:Maximum number of aligned sequences to keep. 设置最多的目标序列匹配数
-num_threads:指定多少个线程运行任务
-outfmt format "7 qacc sacc evalue length pident" :这个是新BLAST+中最拉风的功能了,直接控制输出格式,不用再用parser啦, 7表示带注释行的tab格式的输出,可以自定义要输出哪些内容,用空格分格跟在7的后面,并把所有的输出控制用双引号括起来,其中qacc查询序列的acc,sacc表示目标序列的acc,evalue即是e值,length即是匹配的长度,pident即是序列相同的百分比
-best_hit_overhang, Best Hit algorithm overhang value (recommended value: 0.1)
-best_hit_score_edge,Best Hit algorithm score edge value (recommended value: 0.1)
-task <String, Permissible values: 'blastn' 'blastn-short' 'dc-megablast' 'megablast' 'vecscreen' >
Task to execute Default = `megablast'
megablast 算法
Reference: Zheng Zhang, Scott Schwartz, Lukas Wagner, and Webb Miller (2000), "A greedy algorithm for aligning DNA sequences", J Comput Biol 2000; 7(1-2):203-14.
Zheng Zhang, Scott Schwartz, Lukas Wagner, and Webb Miller. Journal of Computational Biology. February 2000, 7(1-2): 203-214. doi:10.1089/10665270050081478.