






#使用网格搜索来查找最佳的参数组合 from sklearn.model_selection import GridSearchCV param = {"reg_alpha":np.arange(0,5,0.05),"reg_lambda":np.arange(0,2,0.05)} gscv = GridSearchCV(reg,param_grid = param,scoring = "neg_mean_squared_error",cv=cv) #======【TIME WARNING:10~20 mins】======# time0=time() gscv.fit(Xtrain,Ytrain) print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M:%S:%f")) gscv.best_params_ gscv.best_score_ preds = gscv.predict(Xtest) from sklearn.metrics import r2_score,mean_squared_error as MSE r2_score(Ytest,preds) MSE(Ytest,preds) #网格搜索的结果有什么样的含义呢?为什么会出现这样的结果?你相信网格搜索得出的结果吗?试着用数学和你对XGB的理解来解释一下吧



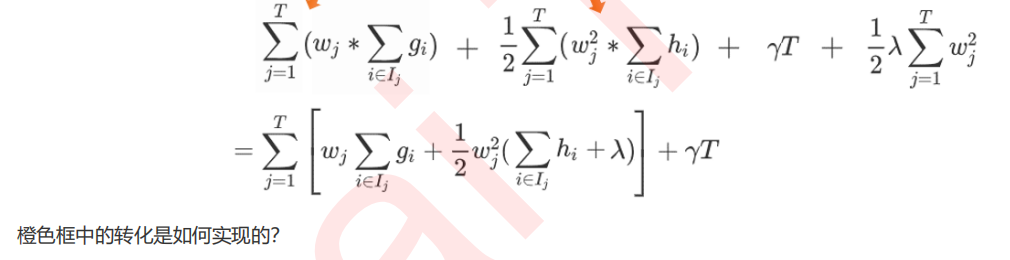
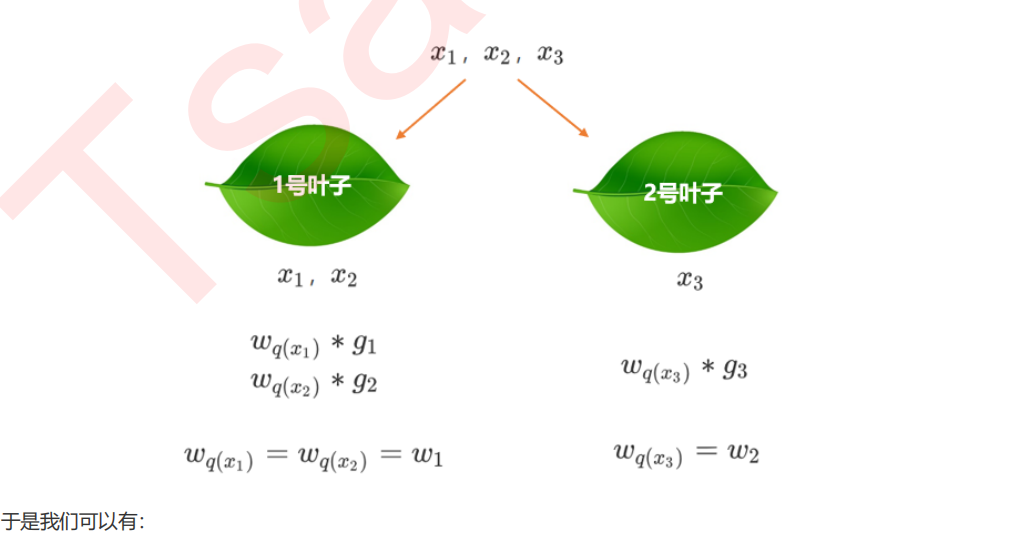
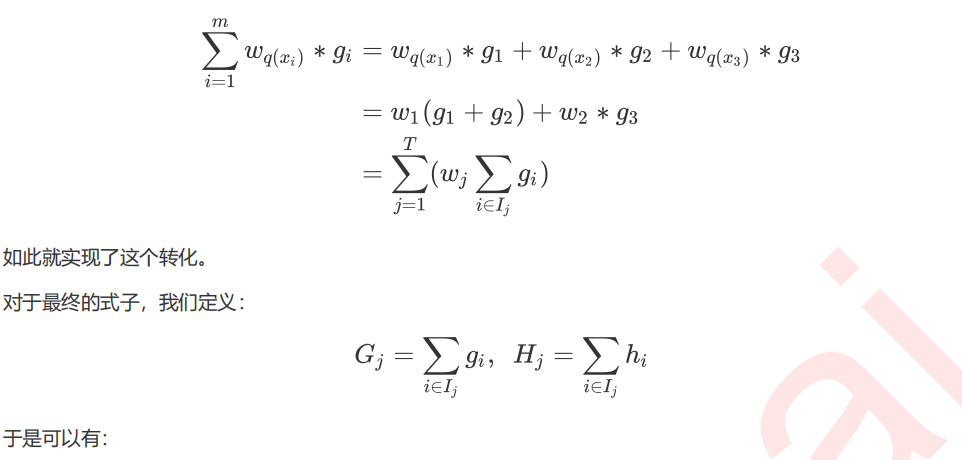


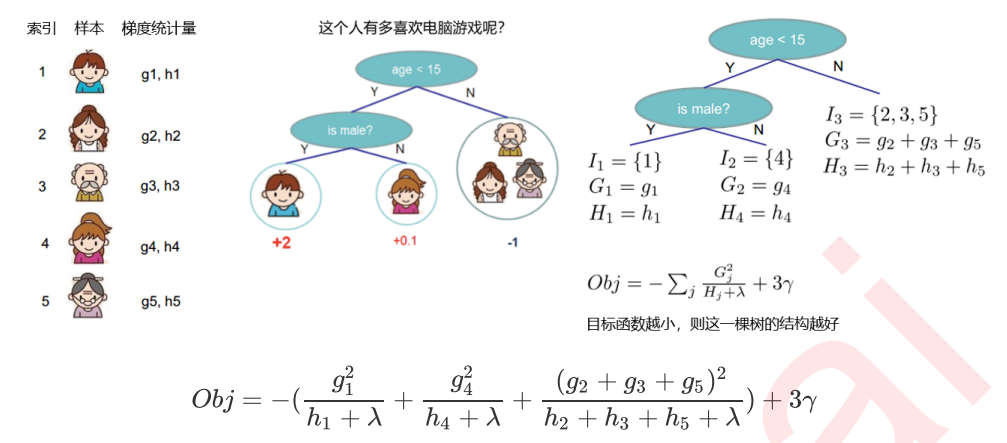



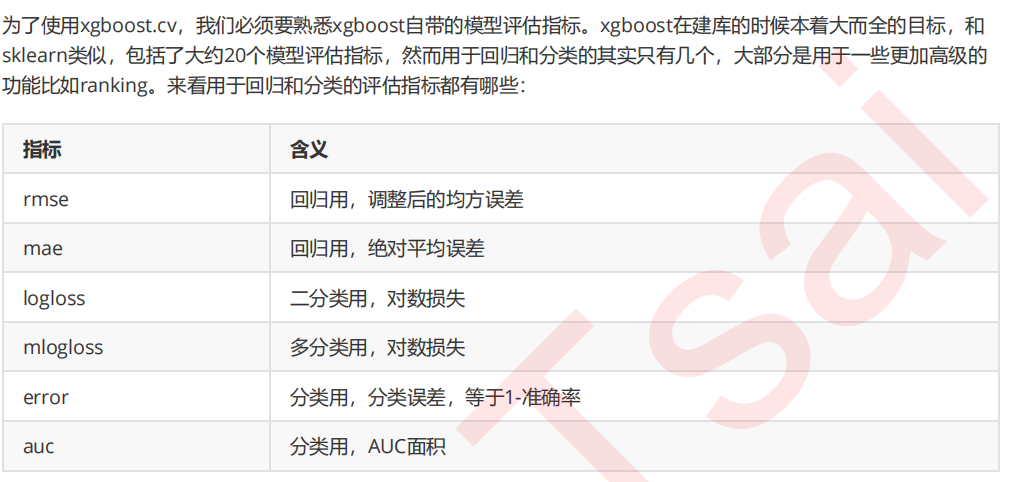
6 寻找最佳分枝:结构分数之差




7 让树停止生长:重要参数gamma


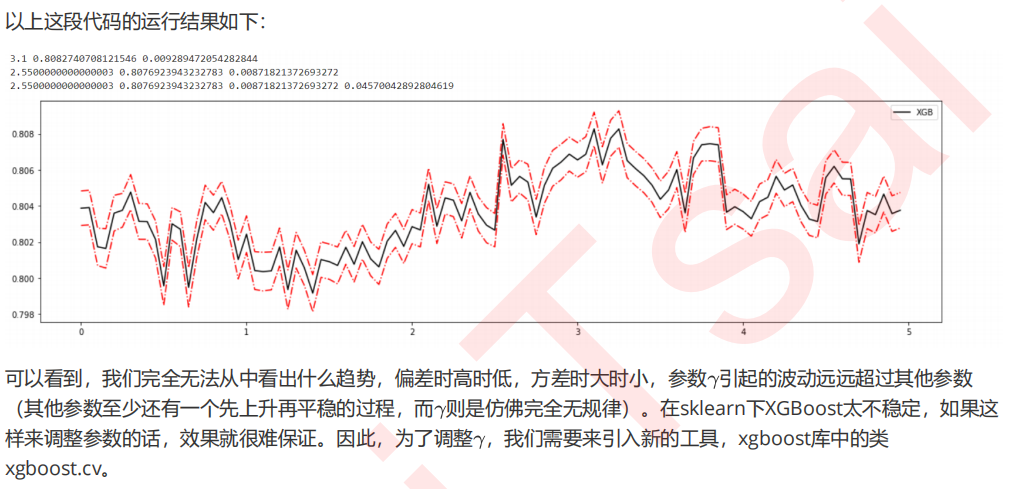
#======【TIME WARNING: 1 min】=======# axisx = np.arange(0,5,0.05) rs = [] var = [] ge = [] for i in axisx: reg = XGBR(n_estimators=180,random_state=420,gamma=i) result = CVS(reg,Xtrain,Ytrain,cv=cv) rs.append(result.mean()) var.append(result.var()) ge.append((1 - result.mean())**2+result.var()) print(axisx[rs.index(max(rs))],max(rs),var[rs.index(max(rs))]) print(axisx[var.index(min(var))],rs[var.index(min(var))],min(var)) print(axisx[ge.index(min(ge))],rs[ge.index(min(ge))],var[ge.index(min(ge))],min(ge)) rs = np.array(rs) var = np.array(var)*0.1 plt.figure(figsize=(20,5)) plt.plot(axisx,rs,c="black",label="XGB") plt.plot(axisx,rs+var,c="red",linestyle='-.') plt.plot(axisx,rs-var,c="red",linestyle='-.') plt.legend() plt.show()

import xgboost as xgb #为了便捷,使用全数据 dfull = xgb.DMatrix(X,y) #设定参数 param1 = {'silent':True,'obj':'reg:linear',"gamma":0} num_round = 180 n_fold=5 #使用类xgb.cv time0 = time() cvresult1 = xgb.cv(param1, dfull, num_round,n_fold) print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M:%S:%f")) #看看类xgb.cv生成了什么结果? cvresult1 plt.figure(figsize=(20,5)) plt.grid() plt.plot(range(1,181),cvresult1.iloc[:,0],c="red",label="train,gamma=0") plt.plot(range(1,181),cvresult1.iloc[:,2],c="orange",label="test,gamma=0") plt.legend() plt.show() #xgboost中回归模型的默认模型评估指标是什么?
param1 = {'silent':True,'obj':'reg:linear',"gamma":0,"eval_metric":"mae"}
cvresult1 = xgb.cv(param1, dfull, num_round,n_fold)
plt.figure(figsize=(20,5))
plt.grid()
plt.plot(range(1,181),cvresult1.iloc[:,0],c="red",label="train,gamma=0")
plt.plot(range(1,181),cvresult1.iloc[:,2],c="orange",label="test,gamma=0")
plt.legend()
plt.show()
#从这个图中,我们可以看出什么?
#怎样从图中观察模型的泛化能力?
#从这个图的角度来说,模型的调参目标是什么?
来看看如果我们调整 ,会发生怎样的变化:
param1 = {'silent':True,'obj':'reg:linear',"gamma":0}
param2 = {'silent':True,'obj':'reg:linear',"gamma":20}
num_round = 180
n_fold=5
time0 = time()
cvresult1 = xgb.cv(param1, dfull, num_round,n_fold)
print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M:%S:%f"))
time0 = time()
cvresult2 = xgb.cv(param2, dfull, num_round,n_fold)
print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M:%S:%f"))
plt.figure(figsize=(20,5))
plt.grid()
plt.plot(range(1,181),cvresult1.iloc[:,0],c="red",label="train,gamma=0")
plt.plot(range(1,181),cvresult1.iloc[:,2],c="orange",label="test,gamma=0")
plt.plot(range(1,181),cvresult2.iloc[:,0],c="green",label="train,gamma=20")
plt.plot(range(1,181),cvresult2.iloc[:,2],c="blue",label="test,gamma=20")
plt.legend()
plt.show()
#从这里,你看出gamma是如何控制过拟合了吗?
试一个分类的例子:
from sklearn.datasets import load_breast_cancer data2 = load_breast_cancer() x2 = data2.data y2 = data2.target dfull2 = xgb.DMatrix(x2,y2) param1 = {'silent':True,'obj':'binary:logistic',"gamma":0,"nfold":5} param2 = {'silent':True,'obj':'binary:logistic',"gamma":2,"nfold":5} num_round = 100 time0 = time() cvresult1 = xgb.cv(param1, dfull2, num_round,metrics=("error")) print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M:%S:%f")) time0 = time() cvresult2 = xgb.cv(param2, dfull2, num_round,metrics=("error")) print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M:%S:%f")) plt.figure(figsize=(20,5)) plt.grid() plt.plot(range(1,101),cvresult1.iloc[:,0],c="red",label="train,gamma=0") plt.plot(range(1,101),cvresult1.iloc[:,2],c="orange",label="test,gamma=0") plt.plot(range(1,101),cvresult2.iloc[:,0],c="green",label="train,gamma=2") plt.plot(range(1,101),cvresult2.iloc[:,2],c="blue",label="test,gamma=2") plt.legend() plt.show()
