python机器学习-乳腺癌细胞挖掘(博主亲自录制视频)
https://study.163.com/course/introduction.htm?courseId=1005269003&utm_campaign=commission&utm_source=cp-400000000398149&utm_medium=share
随机森林
# -*- coding: utf-8 -*-
"""
Created on Tue Sep 4 09:39:29 2018
@author: zhi.li04
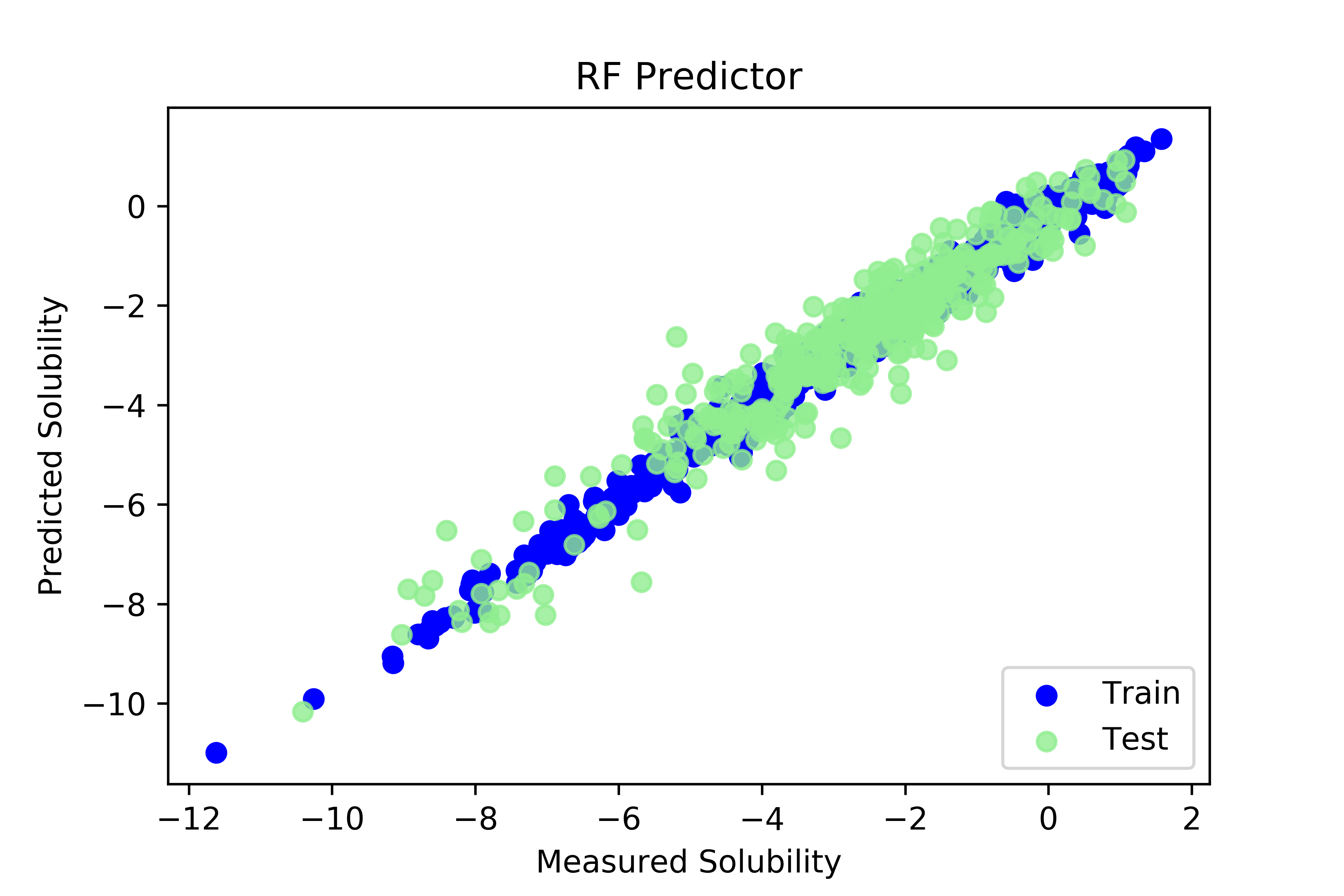
随机森林100棵树
RF RMS 0.6057144333891424
('RF r^2 score', 0.9114913707148344)
随机森林1000棵树
('RF RMS', 0.5891965582822096)
('RF r^2 score', 0.9116131032510899)
"""
from rdkit import Chem, DataStructs
from rdkit.Chem import AllChem
from rdkit.ML.Descriptors import MoleculeDescriptors
from rdkit.Chem import Descriptors
from rdkit.Chem.EState import Fingerprinter
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
from sklearn import cross_validation
from sklearn.metrics import r2_score
from sklearn.ensemble import RandomForestRegressor
from sklearn import gaussian_process
from sklearn.gaussian_process.kernels import Matern, WhiteKernel, ConstantKernel, RBF
#定义描述符计算函数
def get_fps(mol):
calc=MoleculeDescriptors.MolecularDescriptorCalculator([x[0] for x in Descriptors._descList])
ds = np.asarray(calc.CalcDescriptors(mol))
arr=Fingerprinter.FingerprintMol(mol)[0]
return np.append(arr,ds)
#读入数据
data = pd.read_table('smi_sol.dat', sep=' ')
data.to_excel("all_data.xlsx")
#增加结构和描述符属性
data['Mol'] = data['smiles'].apply(Chem.MolFromSmiles)
data['Descriptors'] = data['Mol'].apply(get_fps)
#查看前五行
data.head(5)
#转换为numpy数组
X = np.array(list(data['Descriptors']))
df_x=pd.DataFrame(X)
df_x.to_excel("data.xlsx")
y = data['solubility'].values
df_y=pd.DataFrame(y)
df_y.to_excel("label.xlsx")
st = StandardScaler()
X = st.fit_transform(X)
#划分训练集和测试集
X_train, X_test, y_train, y_test = cross_validation.train_test_split(X, y, test_size=0.3, random_state=42)
'''
#高斯过程回归
kernel=1.0 * RBF(length_scale=1) + WhiteKernel(noise_level=1)
gp = gaussian_process.GaussianProcessRegressor(kernel=kernel,n_restarts_optimizer=0,normalize_y=True)
gp.fit(X_train, y_train)
y_pred, sigma = gp.predict(X_test, return_std=True)
rms = (np.mean((y_test - y_pred)**2))**0.5
#s = np.std(y_test -y_pred)
print ("GP RMS", rms)
print ("GP r^2 score",r2_score(y_test,y_pred))
'''
#随机森林模型
rf = RandomForestRegressor(n_estimators=100)
rf.fit(X_train, y_train)
y_pred = rf.predict(X)
rms = (np.mean((y - y_pred)**2))**0.5
print ("RF RMS", rms)
print ("RF r^2 score",r2_score(y,y_pred))
plt.scatter(y_train,rf.predict(X_train), label = 'Train', c='blue')
plt.title('RF Predictor')
plt.xlabel('Measured Solubility')
plt.ylabel('Predicted Solubility')
plt.scatter(y_test,rf.predict(X_test),c='lightgreen', label='Test', alpha = 0.8)
plt.legend(loc=4)
plt.savefig('RF Predictor.png', dpi=600)
plt.show()
df_validation=pd.DataFrame({"test":y,"predict":y_pred})
df_validation.to_excel("validation.xlsx")
高斯模型
# -*- coding: utf-8 -*-
"""
Created on Tue Sep 4 15:53:57 2018
@author: zhi.li04
默认参数
GP RMS label 2.98575
GP r^2 score -1.26973055888
核参数改为kernel=1.0 * RBF(length_scale=1) + WhiteKernel(noise_level=1)
RMS 0.651688
dtype: float64
GP r^2 score 0.891869923330719
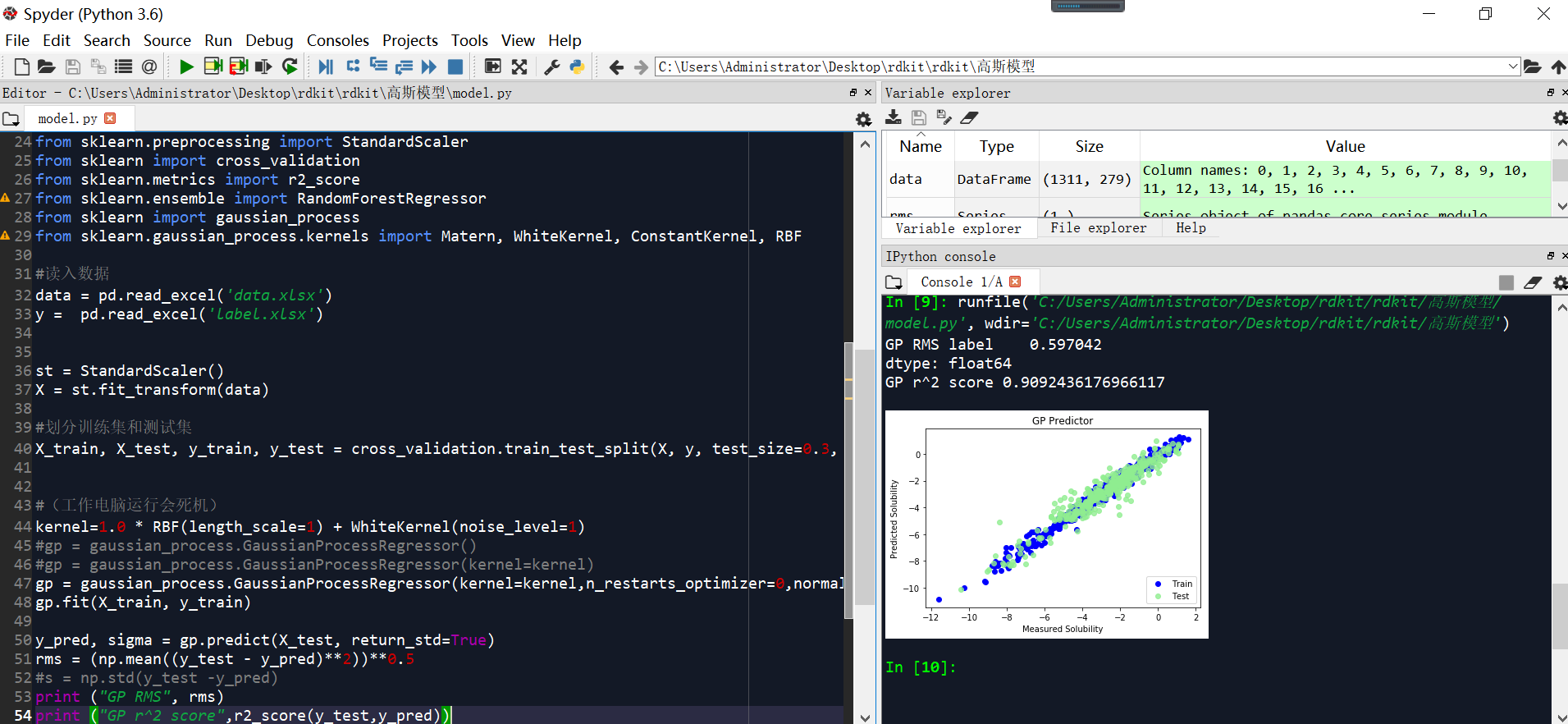
核参数修改,且正态化后
GP RMS label 0.597042
GP r^2 score 0.9092436176966117
"""
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
from sklearn import cross_validation
from sklearn.metrics import r2_score
from sklearn.ensemble import RandomForestRegressor
from sklearn import gaussian_process
from sklearn.gaussian_process.kernels import Matern, WhiteKernel, ConstantKernel, RBF
#读入数据
data = pd.read_excel('data.xlsx')
y = pd.read_excel('label.xlsx')
st = StandardScaler()
X = st.fit_transform(data)
#划分训练集和测试集
X_train, X_test, y_train, y_test = cross_validation.train_test_split(X, y, test_size=0.3, random_state=42)
#(工作电脑运行会死机)
kernel=1.0 * RBF(length_scale=1) + WhiteKernel(noise_level=1)
#gp = gaussian_process.GaussianProcessRegressor()
#gp = gaussian_process.GaussianProcessRegressor(kernel=kernel)
gp = gaussian_process.GaussianProcessRegressor(kernel=kernel,n_restarts_optimizer=0,normalize_y=True)
gp.fit(X_train, y_train)
y_pred, sigma = gp.predict(X_test, return_std=True)
rms = (np.mean((y_test - y_pred)**2))**0.5
#s = np.std(y_test -y_pred)
print ("GP RMS", rms)
print ("GP r^2 score",r2_score(y_test,y_pred))
plt.scatter(y_train,gp.predict(X_train), label = 'Train', c='blue')
plt.title('GP Predictor')
plt.xlabel('Measured Solubility')
plt.ylabel('Predicted Solubility')
plt.scatter(y_test,gp.predict(X_test),c='lightgreen', label='Test', alpha = 0.8)
plt.legend(loc=4)
plt.savefig('GP Predictor.png', dpi=300)
plt.show()
https://study.163.com/provider/400000000398149/index.htm?share=2&shareId=400000000398149( 欢迎关注博主主页,学习python视频资源,还有大量免费python经典文章)
项目合作QQ:231469242