我只讲讲检测部分的模型,后面两样性分类的试验我没有做,这篇论文采用了很多肺结节检测论文都采用的u-net结构,准确地说是具有DPN结构的3D版本的u-net,直接上图。
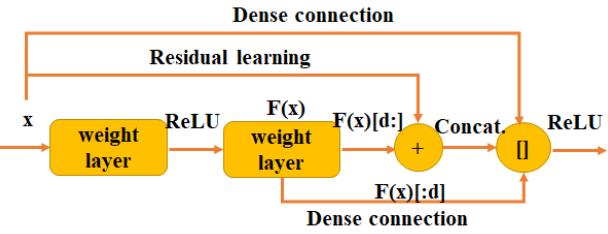
DPN是颜水成老师团队的成果,简单讲就是dense 与 residual的结合,如上图,输入特征图一部分通过residual与输出相加,另一部分与residual的结果再串联,个人觉得这个网络好乱,不简洁的网络都不是好网络,恰好文章中还给出了只采用residual的版本,所以我其实要讲的是这个只有residual的u-net,上图。
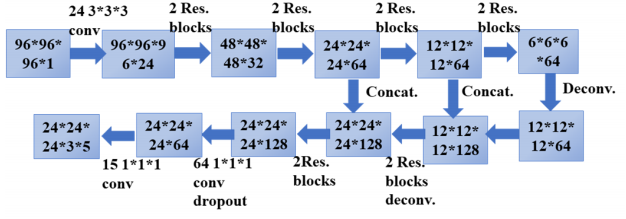
可以看到,输入是96*96*96的立方体,里面包含标记的结节,经过24个3*3*3的卷积核,通道数变为24,然后经过4个stage,尺寸缩减为1/16,接下来是分辨率放大阶段,采用反卷积实现,连续两个阶段都是反卷积后与低层特征串联,然后经过两个卷积操作,通道数变为15,图示中以3*5显示,是为了更清楚地表明,最后输出的proposal中,每个位置有三个,分别采用三种尺寸,设置的三个anchor尺寸是[5,10,20],每个位置预测z,y,x,d,p分别是结节的三维坐标以及直径,置信度。
下面看一下源码,采用pytorch框架。
首先是residual block的设计,位于layers.py文件
class PostRes(nn.Module): def __init__(self, n_in, n_out, stride = 1): super(PostRes, self).__init__() self.conv1 = nn.Conv3d(n_in, n_out, kernel_size = 3, stride = stride, padding = 1) self.bn1 = nn.BatchNorm3d(n_out) self.relu = nn.ReLU(inplace = True) self.conv2 = nn.Conv3d(n_out, n_out, kernel_size = 3, padding = 1) self.bn2 = nn.BatchNorm3d(n_out) if stride != 1 or n_out != n_in: self.shortcut = nn.Sequential( nn.Conv3d(n_in, n_out, kernel_size = 1, stride = stride), nn.BatchNorm3d(n_out)) else: self.shortcut = None def forward(self, x): residual = x if self.shortcut is not None: residual = self.shortcut(x) out = self.conv1(x) out = self.bn1(out) out = self.relu(out) out = self.conv2(out) out = self.bn2(out) out += residual out = self.relu(out) return out
可以看到采用结构与2D的residual基本一致,采用的都是conv-bn-relu,根据步长和输入输出的尺寸,采用identity或1*1卷积作为skip connection。
然后就是网络,位于res18.py文件
class Net(nn.Module):
def __init__(self): super(Net, self).__init__() # The first few layers consumes the most memory, so use simple convolution to save memory. # Call these layers preBlock, i.e., before the residual blocks of later layers. self.preBlock = nn.Sequential( nn.Conv3d(1, 24, kernel_size = 3, padding = 1), nn.BatchNorm3d(24), nn.ReLU(inplace = True), nn.Conv3d(24, 24, kernel_size = 3, padding = 1), nn.BatchNorm3d(24), nn.ReLU(inplace = True)) # 3 poolings, each pooling downsamples the feature map by a factor 2. # 3 groups of blocks. The first block of each group has one pooling. num_blocks_forw = [2,2,3,3] num_blocks_back = [3,3] self.featureNum_forw = [24,32,64,64,64] self.featureNum_back = [128,64,64] for i in range(len(num_blocks_forw)): blocks = [] for j in range(num_blocks_forw[i]): if j == 0: blocks.append(PostRes(self.featureNum_forw[i], self.featureNum_forw[i+1])) else: blocks.append(PostRes(self.featureNum_forw[i+1], self.featureNum_forw[i+1])) setattr(self, 'forw' + str(i + 1), nn.Sequential(*blocks)) for i in range(len(num_blocks_back)): blocks = [] for j in range(num_blocks_back[i]): if j == 0: if i==0: addition = 3 else: addition = 0 blocks.append(PostRes(self.featureNum_back[i+1]+self.featureNum_forw[i+2]+addition, self.featureNum_back[i])) else: blocks.append(PostRes(self.featureNum_back[i], self.featureNum_back[i])) setattr(self, 'back' + str(i + 2), nn.Sequential(*blocks)) self.maxpool1 = nn.MaxPool3d(kernel_size=2,stride=2,return_indices =True) self.maxpool2 = nn.MaxPool3d(kernel_size=2,stride=2,return_indices =True) self.maxpool3 = nn.MaxPool3d(kernel_size=2,stride=2,return_indices =True) self.maxpool4 = nn.MaxPool3d(kernel_size=2,stride=2,return_indices =True) self.unmaxpool1 = nn.MaxUnpool3d(kernel_size=2,stride=2) self.unmaxpool2 = nn.MaxUnpool3d(kernel_size=2,stride=2) self.path1 = nn.Sequential( nn.ConvTranspose3d(64, 64, kernel_size = 2, stride = 2), nn.BatchNorm3d(64), nn.ReLU(inplace = True)) self.path2 = nn.Sequential( nn.ConvTranspose3d(64, 64, kernel_size = 2, stride = 2), nn.BatchNorm3d(64*k), nn.ReLU(inplace = True)) self.drop = nn.Dropout3d(p = 0.5, inplace = False) self.output = nn.Sequential(nn.Conv3d(self.featureNum_back[0], 64, kernel_size = 1), nn.ReLU(), #nn.Dropout3d(p = 0.3), nn.Conv3d(64, 5 * len(config['anchors']), kernel_size = 1)) def forward(self, x, coord): out = self.preBlock(x)#16 out_pool,indices0 = self.maxpool1(out) out1 = self.forw1(out_pool)#32 out1_pool,indices1 = self.maxpool2(out1) out2 = self.forw2(out1_pool)#64 #out2 = self.drop(out2) out2_pool,indices2 = self.maxpool3(out2) out3 = self.forw3(out2_pool)#64 out3_pool,indices3 = self.maxpool4(out3) out4 = self.forw4(out3_pool)#64 #out4 = self.drop(out4) rev3 = self.path1(out4) comb3 = self.back3(torch.cat((rev3, out3), 1))#64+64 #comb3 = self.drop(comb3) rev2 = self.path2(comb3) comb2 = self.back2(torch.cat((rev2, out2,coord), 1))#128 comb2 = self.drop(comb2) out = self.output(comb2) size = out.size() out = out.view(out.size(0), out.size(1), -1) #out = out.transpose(1, 4).transpose(1, 2).transpose(2, 3).contiguous() out = out.transpose(1, 2).contiguous().view(size[0], size[2], size[3], size[4], len(config['anchors']), 5) #out = out.view(-1, 5) return out
看代码的时候有个地方比较绕,就是forw模块和back模块的迭代实现,个人觉得还不如直接一个模块一个模块地写出来,虽然多点代码,但比较清晰。还有就是path模块,其实就是反卷积模块。
网络结构就是这些,其实难点在loss的定义,以及标签的映射,下面来看一下loss的定义,标签映射以及数据增强部分待到(中)(下)部再讲。
loss的定义采用的也是pytorch网络的定义,位于layers.py文件。
上代码。
class Loss(nn.Module): def __init__(self, num_hard = 0): super(Loss, self).__init__() self.sigmoid = nn.Sigmoid() self.classify_loss = nn.BCELoss() #二分类交叉熵损失 self.regress_loss = nn.SmoothL1Loss() #平滑L1损失 self.num_hard = num_hard #hardming 数目 def forward(self, output, labels, train = True): batch_size = labels.size(0) #标签的第0维度,样本数 output = output.view(-1, 5) #将输出维度调整,以anchor为第二维度 labels = labels.view(-1, 5) #将标签维度对应调整,同上 pos_idcs = labels[:, 0] > 0.5 #对标签进行筛选,输出为索引,示例[1,2,5] pos_idcs = pos_idcs.unsqueeze(1).expand(pos_idcs.size(0), 5) #对索引维度扩展,重复5次,示例[[1,1,1,1,1],[2,2,2,2,2],[5,5,5,5,5]] pos_output = output[pos_idcs].view(-1, 5) #筛选出与正标签对应的输出 pos_labels = labels[pos_idcs].view(-1, 5) #筛选出正标签 neg_idcs = labels[:, 0] < -0.5 #同上,筛选负标签索引,此处为负值 neg_output = output[:, 0][neg_idcs] #注意,此处与上面不同,负标签只考虑置信度即可,因为位置及直径不计入损失,没有意义 neg_labels = labels[:, 0][neg_idcs] if self.num_hard > 0 and train:#判断是否定义了,hardmining neg_output, neg_labels = hard_mining(neg_output, neg_labels, self.num_hard * batch_size) #只选择置信度较高的负样本作计算,对于易于分类的负样本,都是虾兵蟹将,不足虑 neg_prob = self.sigmoid(neg_output)#对负样本输出进行sigmoid处理,生成0~1之间的值,符合置信度的范围,可能大家要问输出不就是0~1吗,这里网络最后没有用sigmoid激活函数,所以最后输出应该是没有范围的, #这里我也比较不解,直接在网络中加入sigmoid不就行了 #classify_loss = self.classify_loss( # torch.cat((pos_prob, neg_prob), 0), # torch.cat((pos_labels[:, 0], neg_labels + 1), 0)) if len(pos_output)>0: pos_prob = self.sigmoid(pos_output[:, 0]) #对正样本进行sigmoid处理 pz, ph, pw, pd = pos_output[:, 1], pos_output[:, 2], pos_output[:, 3], pos_output[:, 4] #依次输出z,h,w,d以便与标签结合求损失 lz, lh, lw, ld = pos_labels[:, 1], pos_labels[:, 2], pos_labels[:, 3], pos_labels[:, 4] #依次输出z,h,w,d以便与输出结合求损失 regress_losses = [ #回归损失 self.regress_loss(pz, lz), self.regress_loss(ph, lh), self.regress_loss(pw, lw), self.regress_loss(pd, ld)] regress_losses_data = [l.data[0] for l in regress_losses] classify_loss = 0.5 * self.classify_loss( #对正样本和负样本分别求分类损失 pos_prob, pos_labels[:, 0]) + 0.5 * self.classify_loss( neg_prob, neg_labels + 1) pos_correct = (pos_prob.data >= 0.5).sum() #那些输出确实大于0.5的正样本是正确预测的正样本 pos_total = len(pos_prob) #正样本总数 else: #如果没有正标签,由于负标签又不用计算回归损失,于是回归损失就置零了,分类损失只计算负标签的分类损失 regress_losses = [0,0,0,0] classify_loss = 0.5 * self.classify_loss( neg_prob, neg_labels + 1) pos_correct = 0 #此时没有正样本或正标签 pos_total = 0 #总数也为0 regress_losses_data = [0,0,0,0] classify_loss_data = classify_loss.data[0] #loss = classify_loss#pytorch 0.4 loss = classify_loss.clone() for regress_loss in regress_losses: #将回归损失与分类损失相加,求出总损失(标量) loss += regress_loss neg_correct = (neg_prob.data < 0.5).sum() #那些输出确实低于0.5的负样本是正确预测的负样本 neg_total = len(neg_prob) #负样本总数 return [loss, classify_loss_data] + regress_losses_data + [pos_correct, pos_total, neg_correct, neg_total]
对于损失的解释都在代码旁边的注释了,只是有一点不大明白,求负样本损失的时候为何要把置信度加1?,应该是负标签在打标签的时候置为-1了,由此又想到一个问题,那些既非正也非负的样本的置信度是如何设置的,应该不是随机设置的,难道设为0了?
在(中)里面,我想把标签映射以及数据增强,讲一下,奈何自己还不太懂,等等吧,如果(中)完成,在(下)里简单说一说训练以及验证,以及测试,这些都完成,那么deeplung笔记三部曲连在一起就完整了。