第七章、支持向量机SVM(非线性)
1.逻辑回归支持向量机
代价函数:
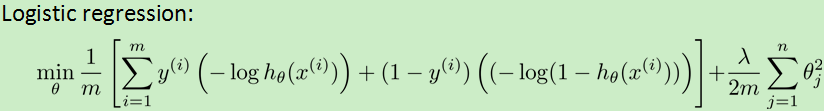
用cost1(z)代替![]() ,cost0(z)代替
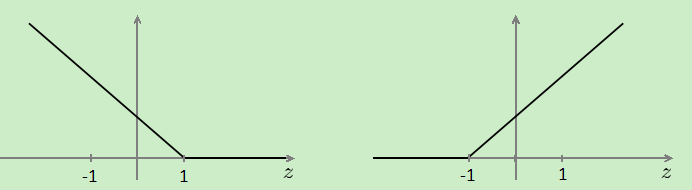
,cost0(z)代替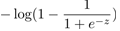 ,两个函数的图像:
,两个函数的图像:
去掉m,用C代替lambda,得支持向量机的算法:
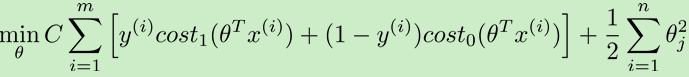
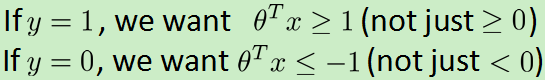
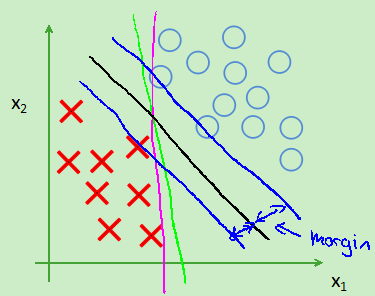
支持向量机的间距(大间隔分类):因为theta' *X>=1而不是0,所以会选择一条离数据最远的一条线,如图:会选择那条黑线
2.支持向量机能实现大间隔分类的原因:
设Ax+By=0表示一条线段,则(A,B)可以表示一条向量,并且和直线垂直。向量相乘可以等化为投影(有正负)乘以向量长度。p(i)表示X(i)在theta上的投影。原式可以转化为如下:
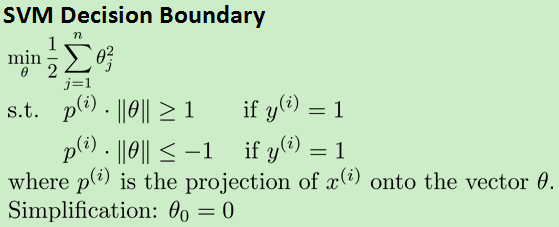
所以会尽可能让p(i)大,就会实现大间隔分类。
3.核函数kernel:处理更加复杂的非线性问题
对每一个数据做一个等价代换:
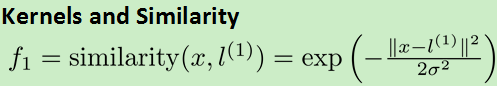
核函数性质:

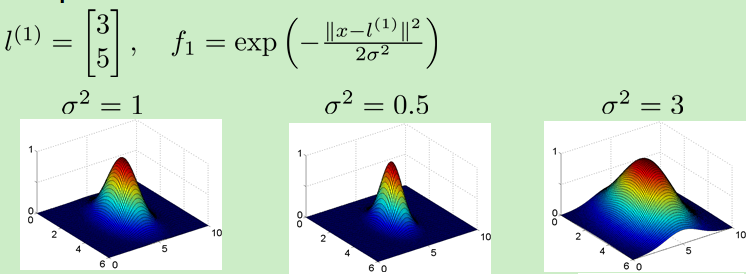
![]() 的影响:越大越平缓
的影响:越大越平缓
预测函数:
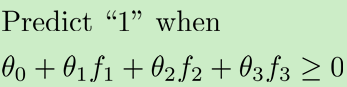
对于一个问题,![]() ?
?
用每个数据集:
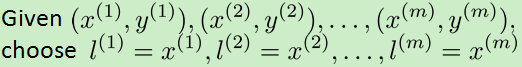
支持向量机的核函数求theta的h函数和cost函数:
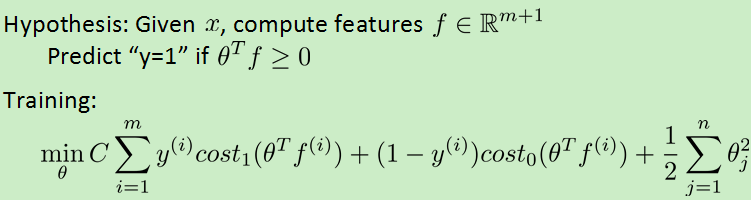
注意上图中的正则化项在支持向量机中一般用theta' *m*theta(m为数据集个数),目的是提高效率。
4.支持向量机的核函数的高偏差和高方差:
C较大:低偏差高方差
C较小:高偏差低方差![]() 较大:图像较平缓,高偏差低方差
较大:图像较平缓,高偏差低方差![]() 较小:图像较陡峭,低偏差高方差
较小:图像较陡峭,低偏差高方差
5.在octave中使用自带的支持向量机的函数,需要提供一个函数实现(核函数):

在使用高斯函数前,一定要归一化,否则差距太大,可能会忽略掉小的特征值。
6.多分类:一对多,选最大的,和逻辑回归类似。
7.分类问题的选择
- 如果特征n相比数据集m较大,则一般不需要非常复杂的边界,用支持向量机的线性核函数(不使用核函数)或逻辑回归来形成一个简单地边界(大约:n=10000,m=10-1000)
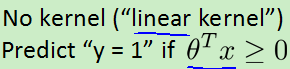
- 相反如果数据集较多,则想要一个较为复杂曲线的边界,用前面提到的支持向量机的高斯核函数(大约:n=1-1000,m=10-10000)

- 当m太大时,高斯核函数运行会很慢,一般会手动增加特征,并用线性核函数(不使用核函数)或逻辑回归来处理(m=50000+)
- 神经网络求解也很慢,优化的SVM会比它快,并且存在局部最优解而非整体最优解,能处理非常复杂的问题。
代码段1:支持向量机的线性核函数和高斯核函数(不是自己写的)mlclass-ex6-007
1.线性核函数
function sim = linearKernel(x1, x2) x1 = x1(:); x2 = x2(:); sim = x1' * x2; % 不知道为什么? end
2.高斯核函数
function sim = gaussianKernel(x1, x2, sigma) x1 = x1(:); x2 = x2(:); sim = 0; x=x1-x2; sim=exp(-(x' * x)/(2*sigma*sigma) ); end
3.高斯核函数选取![]() 和C
和C
function [C, sigma] = dataset3Params(X, y, Xval, yval) C = 1; sigma = 0.3; value=[0.01 0.03 0.1 0.3 1 3 10 30]'; error=2; for i=1:size(value,1) tC=value(i); for j=1:size(value,1) tsigma=value(j); model= svmTrain(X, y, tC, @(x1, x2) gaussianKernel(x1, x2, tsigma)); %支持向量机算法 predictions = svmPredict(model, Xval); %支持向量机预测算法 terror=mean(double(predictions ~= yval)); if terror<error error=terror; C=tC; sigma=tsigma; end; end; end; end
4.svmTrain,visualizeBoundaryLinear,visualizeBoundary,svmPredict(自带)

function [model] = svmTrain(X, Y, C, kernelFunction, ... tol, max_passes) %SVMTRAIN Trains an SVM classifier using a simplified version of the SMO %algorithm. % [model] = SVMTRAIN(X, Y, C, kernelFunction, tol, max_passes) trains an % SVM classifier and returns trained model. X is the matrix of training % examples. Each row is a training example, and the jth column holds the % jth feature. Y is a column matrix containing 1 for positive examples % and 0 for negative examples. C is the standard SVM regularization % parameter. tol is a tolerance value used for determining equality of % floating point numbers. max_passes controls the number of iterations % over the dataset (without changes to alpha) before the algorithm quits. % % Note: This is a simplified version of the SMO algorithm for training % SVMs. In practice, if you want to train an SVM classifier, we % recommend using an optimized package such as: % % LIBSVM (http://www.csie.ntu.edu.tw/~cjlin/libsvm/) % SVMLight (http://svmlight.joachims.org/) % % if ~exist('tol', 'var') || isempty(tol) tol = 1e-3; end if ~exist('max_passes', 'var') || isempty(max_passes) max_passes = 5; end % Data parameters m = size(X, 1); n = size(X, 2); % Map 0 to -1 Y(Y==0) = -1; % Variables alphas = zeros(m, 1); b = 0; E = zeros(m, 1); passes = 0; eta = 0; L = 0; H = 0; % Pre-compute the Kernel Matrix since our dataset is small % (in practice, optimized SVM packages that handle large datasets % gracefully will _not_ do this) % % We have implemented optimized vectorized version of the Kernels here so % that the svm training will run faster. if strcmp(func2str(kernelFunction), 'linearKernel') % Vectorized computation for the Linear Kernel % This is equivalent to computing the kernel on every pair of examples K = X*X'; elseif strfind(func2str(kernelFunction), 'gaussianKernel') % Vectorized RBF Kernel % This is equivalent to computing the kernel on every pair of examples X2 = sum(X.^2, 2); K = bsxfun(@plus, X2, bsxfun(@plus, X2', - 2 * (X * X'))); K = kernelFunction(1, 0) .^ K; else % Pre-compute the Kernel Matrix % The following can be slow due to the lack of vectorization K = zeros(m); for i = 1:m for j = i:m K(i,j) = kernelFunction(X(i,:)', X(j,:)'); K(j,i) = K(i,j); %the matrix is symmetric end end end % Train fprintf(' Training ...'); dots = 12; while passes < max_passes, num_changed_alphas = 0; for i = 1:m, % Calculate Ei = f(x(i)) - y(i) using (2). % E(i) = b + sum (X(i, :) * (repmat(alphas.*Y,1,n).*X)') - Y(i); E(i) = b + sum (alphas.*Y.*K(:,i)) - Y(i); if ((Y(i)*E(i) < -tol && alphas(i) < C) || (Y(i)*E(i) > tol && alphas(i) > 0)), % In practice, there are many heuristics one can use to select % the i and j. In this simplified code, we select them randomly. j = ceil(m * rand()); while j == i, % Make sure i eq j j = ceil(m * rand()); end % Calculate Ej = f(x(j)) - y(j) using (2). E(j) = b + sum (alphas.*Y.*K(:,j)) - Y(j); % Save old alphas alpha_i_old = alphas(i); alpha_j_old = alphas(j); % Compute L and H by (10) or (11). if (Y(i) == Y(j)), L = max(0, alphas(j) + alphas(i) - C); H = min(C, alphas(j) + alphas(i)); else L = max(0, alphas(j) - alphas(i)); H = min(C, C + alphas(j) - alphas(i)); end if (L == H), % continue to next i. continue; end % Compute eta by (14). eta = 2 * K(i,j) - K(i,i) - K(j,j); if (eta >= 0), % continue to next i. continue; end % Compute and clip new value for alpha j using (12) and (15). alphas(j) = alphas(j) - (Y(j) * (E(i) - E(j))) / eta; % Clip alphas(j) = min (H, alphas(j)); alphas(j) = max (L, alphas(j)); % Check if change in alpha is significant if (abs(alphas(j) - alpha_j_old) < tol), % continue to next i. % replace anyway alphas(j) = alpha_j_old; continue; end % Determine value for alpha i using (16). alphas(i) = alphas(i) + Y(i)*Y(j)*(alpha_j_old - alphas(j)); % Compute b1 and b2 using (17) and (18) respectively. b1 = b - E(i) ... - Y(i) * (alphas(i) - alpha_i_old) * K(i,j)' ... - Y(j) * (alphas(j) - alpha_j_old) * K(i,j)'; b2 = b - E(j) ... - Y(i) * (alphas(i) - alpha_i_old) * K(i,j)' ... - Y(j) * (alphas(j) - alpha_j_old) * K(j,j)'; % Compute b by (19). if (0 < alphas(i) && alphas(i) < C), b = b1; elseif (0 < alphas(j) && alphas(j) < C), b = b2; else b = (b1+b2)/2; end num_changed_alphas = num_changed_alphas + 1; end end if (num_changed_alphas == 0), passes = passes + 1; else passes = 0; end fprintf('.'); dots = dots + 1; if dots > 78 dots = 0; fprintf(' '); end if exist('OCTAVE_VERSION') fflush(stdout); end end fprintf(' Done! '); % Save the model idx = alphas > 0; model.X= X(idx,:); model.y= Y(idx); model.kernelFunction = kernelFunction; model.b= b; model.alphas= alphas(idx); model.w = ((alphas.*Y)'*X)'; end

function visualizeBoundaryLinear(X, y, model) %VISUALIZEBOUNDARYLINEAR plots a linear decision boundary learned by the %SVM % VISUALIZEBOUNDARYLINEAR(X, y, model) plots a linear decision boundary % learned by the SVM and overlays the data on it w = model.w; b = model.b; xp = linspace(min(X(:,1)), max(X(:,1)), 100); yp = - (w(1)*xp + b)/w(2); plotData(X, y); hold on; plot(xp, yp, '-b'); hold off end

function visualizeBoundary(X, y, model, varargin) %VISUALIZEBOUNDARY plots a non-linear decision boundary learned by the SVM % VISUALIZEBOUNDARYLINEAR(X, y, model) plots a non-linear decision % boundary learned by the SVM and overlays the data on it % Plot the training data on top of the boundary plotData(X, y) % Make classification predictions over a grid of values x1plot = linspace(min(X(:,1)), max(X(:,1)), 100)'; x2plot = linspace(min(X(:,2)), max(X(:,2)), 100)'; [X1, X2] = meshgrid(x1plot, x2plot); vals = zeros(size(X1)); for i = 1:size(X1, 2) this_X = [X1(:, i), X2(:, i)]; vals(:, i) = svmPredict(model, this_X); end % Plot the SVM boundary hold on contour(X1, X2, vals, [0 0], 'Color', 'b'); hold off; end

function pred = svmPredict(model, X) %SVMPREDICT returns a vector of predictions using a trained SVM model %(svmTrain). % pred = SVMPREDICT(model, X) returns a vector of predictions using a % trained SVM model (svmTrain). X is a mxn matrix where there each % example is a row. model is a svm model returned from svmTrain. % predictions pred is a m x 1 column of predictions of {0, 1} values. % % Check if we are getting a column vector, if so, then assume that we only % need to do prediction for a single example if (size(X, 2) == 1) % Examples should be in rows X = X'; end % Dataset m = size(X, 1); p = zeros(m, 1); pred = zeros(m, 1); if strcmp(func2str(model.kernelFunction), 'linearKernel') % We can use the weights and bias directly if working with the % linear kernel p = X * model.w + model.b; elseif strfind(func2str(model.kernelFunction), 'gaussianKernel') % Vectorized RBF Kernel % This is equivalent to computing the kernel on every pair of examples X1 = sum(X.^2, 2); X2 = sum(model.X.^2, 2)'; K = bsxfun(@plus, X1, bsxfun(@plus, X2, - 2 * X * model.X')); K = model.kernelFunction(1, 0) .^ K; K = bsxfun(@times, model.y', K); K = bsxfun(@times, model.alphas', K); p = sum(K, 2); else % Other Non-linear kernel for i = 1:m prediction = 0; for j = 1:size(model.X, 1) prediction = prediction + ... model.alphas(j) * model.y(j) * ... model.kernelFunction(X(i,:)', model.X(j,:)'); end p(i) = prediction + model.b; end end % Convert predictions into 0 / 1 pred(p >= 0) = 1; pred(p < 0) = 0; end
5.整体代码
clear ; close all; clc load('ex6data1.mat'); % Plot training data %plotData(X, y); %C = 10; %model = svmTrain(X, y, C, @linearKernel, 1e-3, 20); %visualizeBoundaryLinear(X, y, model); x1 = [1 2 1]; x2 = [0 4 -1]; sigma = 2; sim = gaussianKernel(x1, x2, sigma); clear ; close all; clc load('ex6data2.mat'); %plotData(X, y); C = 1; sigma = 0.1; %model= svmTrain(X, y, C, @(x1, x2) gaussianKernel(x1, x2, sigma)); %visualizeBoundary(X, y, model); clear ; close all; clc load('ex6data3.mat'); %plotData(X, y); [C, sigma] = dataset3Params(X, y, Xval, yval); model= svmTrain(X, y, C, @(x1, x2) gaussianKernel(x1, x2, sigma)); visualizeBoundary(X, y, model);
代码段2:垃圾邮件分类 mlclass-ex6-007
字符串处理:将邮件进行预处理,只保留单词。
选择出现频率最高的单词,然后形成向量。
