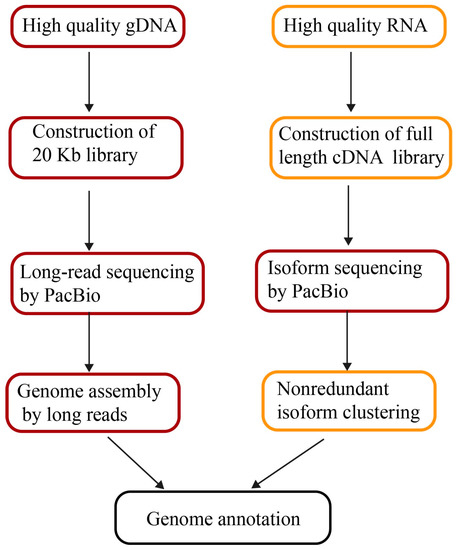
Genome Sequencing and Assembly by Long Reads in Plants 植物基因组的长read测序和组装
1
College of Agronomy, Shenyang Agricultural University, 120 Dongling Road, Shenyang 110866, China
2
College of Bioscience and Biotechnology, Shenyang Agricultural University, 120 Dongling Road, Shenyang 110866, China
3
School of Agriculture and Biology, Shanghai Jiao Tong University, 800 Dong Chuan Road, Shanghai 200240, China
*
Authors to whom correspondence should be addressed.
Genes 2018, 9(1), 6; https://doi.org/10.3390/genes9010006
Received: 20 November 2017 / Revised: 18 December 2017 / Accepted: 18 December 2017 / Published: 28 December 2017
(This article belongs to the Special Issue Plant Genomics and Epigenomics for Trait Improvement)
Plant genomes generated by Sanger and Next Generation Sequencing (NGS) have provided insight into species diversity and evolution. However, Sanger sequencing is limited in its applications due to high cost, labor intensity, and low throughput, while NGS reads are too short to resolve abundant repeats and polyploidy, leading to incomplete or ambiguous assemblies. The advent and improvement of long-read sequencing by Third Generation Sequencing (TGS) methods such as PacBio and Nanopore have shown promise in producing high-quality assemblies for complex genomes. Here, we review the development of sequencing, introducing the application as well as considerations of experimental design in TGS of plant genomes. We also introduce recent revolutionary scaffolding technologies including BioNano, Hi-C, and 10× Genomics. We expect that the informative guidance for genome sequencing and assembly by long reads will benefit the initiation of scientists’ projects.
由桑格和下一代测序(NGS)生成的植物基因组为物种多样性和进化提供了深入的认识。
然而,Sanger测序由于成本高、劳动强度大、通量低,应用受到限制,而NGS reads太短,无法解决大量重复和多倍体,导致组装不完整或不明确。
第三代测序(TGS)方法如PacBio和Nanopore的出现和改进为生产高质量的复杂基因组组件带来了希望。
本文综述了测序技术的发展,介绍了在植物基因组TGS中的应用以及实验设计的考虑。
我们还介绍了最近革命性的脚手架技术,包括BioNano、Hi-C和10×Genomics。
我们期望通过长读对基因组测序和组装的信息指导将有利于启动科学家的项目。
Keywords: genome assembly; long reads; Sanger sequencing; Next Generation Sequencing; Third Generation Sequencing
▼ Show Figures
This is an open access article distributed under the Creative Commons Attribution License which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited